Search Count: 6,031
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: 1PE, CL, PEG, PGE |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PG4, CA, MES, 1PE |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (Orthorhombic P Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, 1PE |
 |
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: GOL, PGE |
 |
Crystal Structure Of A Malonate Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: MLI, CL, NA |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P61 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, CL, PG4 |
 |
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis In Complex With Pyrophosphate
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PPV |
 |
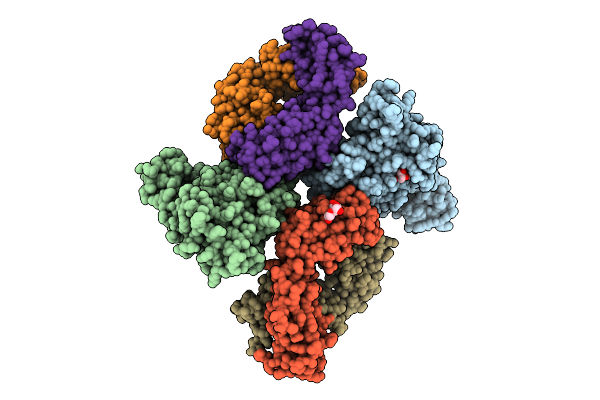
Organism: Archaeoglobus fulgidus dsm 4304, Pyrococcus furiosus dsm 3638, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
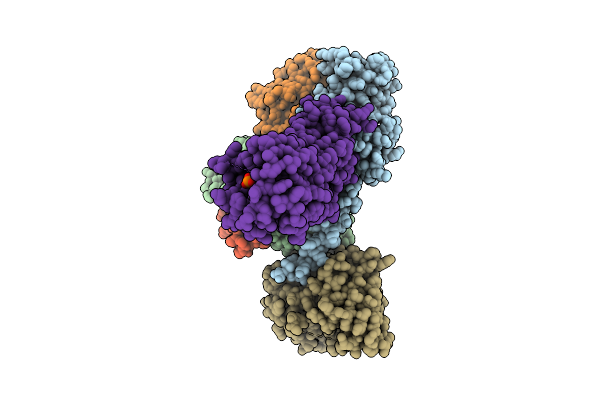
Organism: Rift valley fever virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: GOL, PEG |
 |
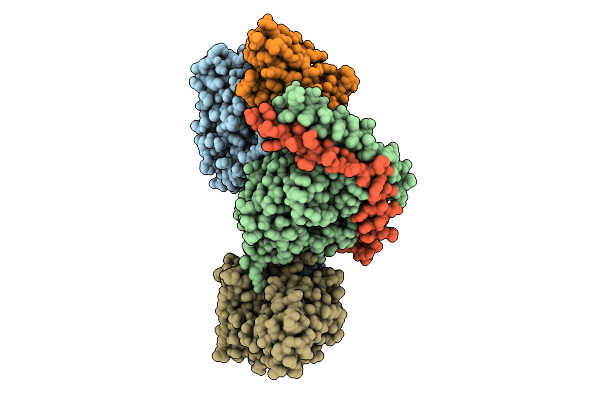
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1L69 |
 |
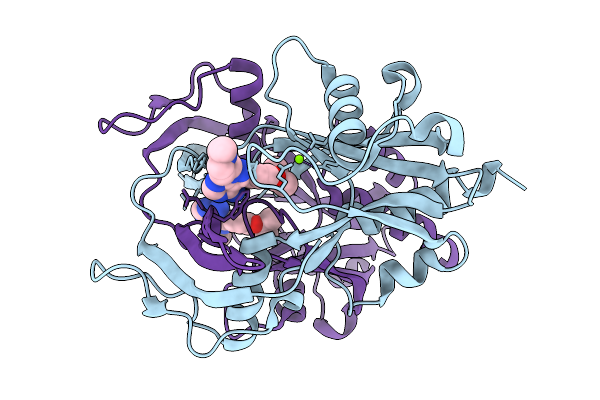
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: S1P |
 |
Crystal Structure Of Human Nudt14 In Complex With A Potent Inhibitor (Ma-955-10)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: A1ITN, MG |
 |
Organism: Colletotrichum orbiculare
Method: SOLUTION NMR Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
 |
Carbohydrate-Binding Module 32 Of Lnbb From Bifidobacterium Bifidum, Ligand Free Form, Multiple Small-Wedge Data Set
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of Human Complement C1S Cub Domain In Complex With Ray121
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: CA |
 |
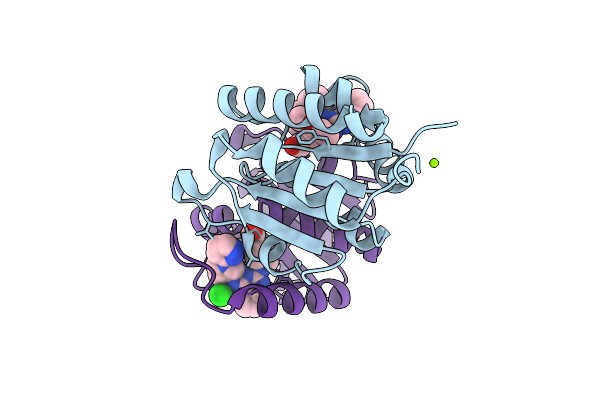
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1IZR |
 |
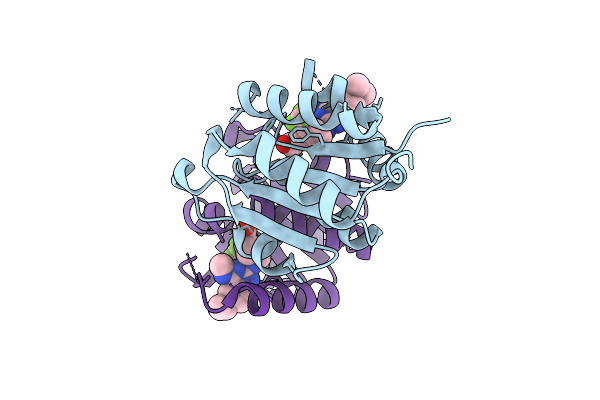
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: MG, A1IZW, CL |
 |
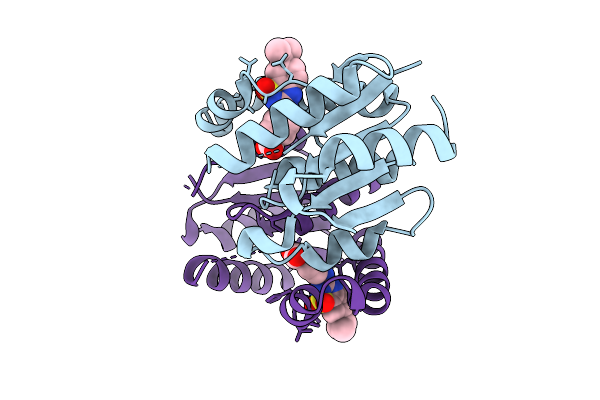
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1IZX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1I0K |

