Search Count: 24,992
 |
Mycobacterium Tuberculosis Relbe1 Toxin-Antitoxin System; Rv1247C (Relb1 Antitoxin), Rv1246C (Rele1 Toxin)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TOXIN Ligands: CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: NIO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
 |
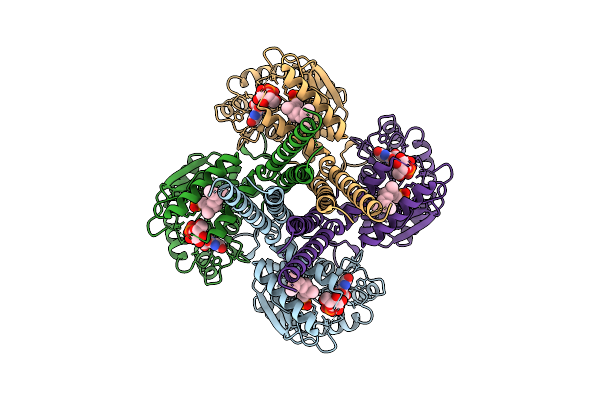
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
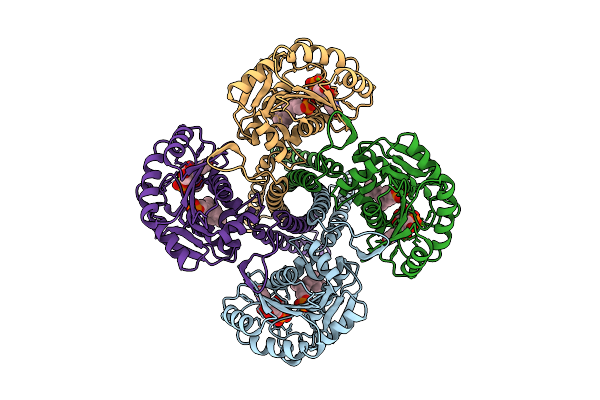
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
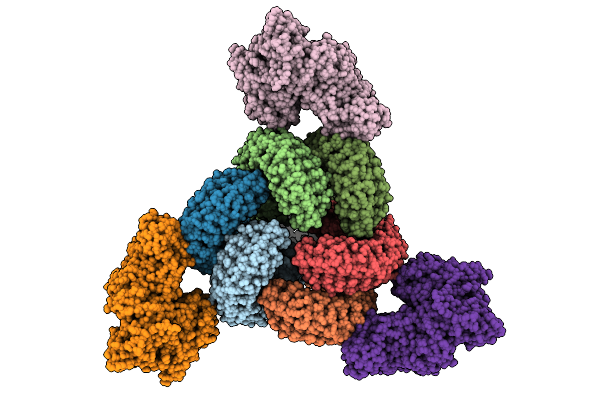
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
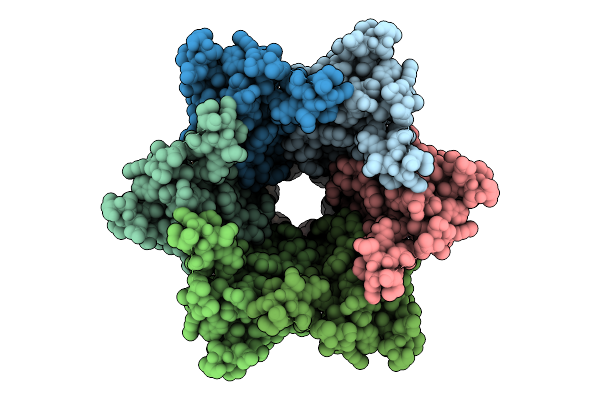
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
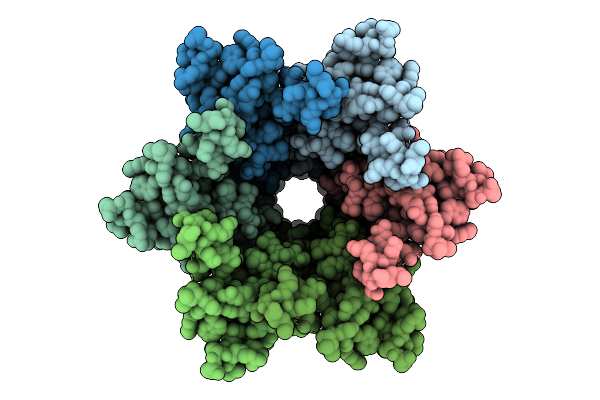
Connexin-32 (Cx32) Gap Junction Channel In Popc-Containing Nanodiscs In The Absence Of Chs
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
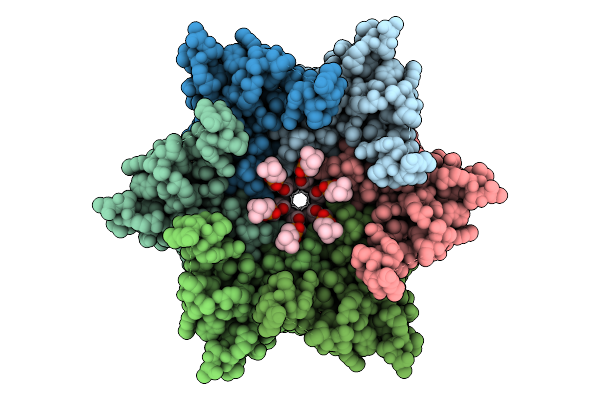
Connexin-32 (Cx32) W3S Mutant Gap Junction Channel In Popc-Containing Msp2N2 Nanodiscs
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
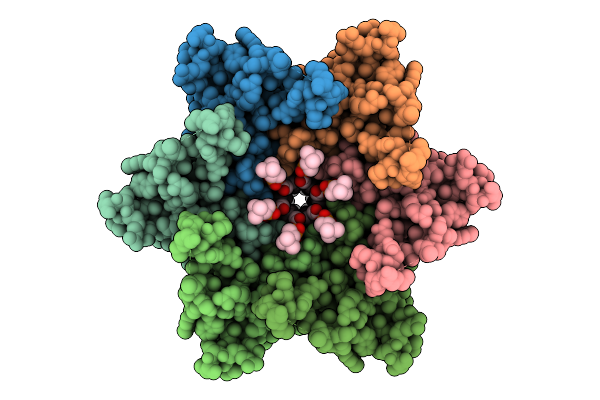
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: POV, CLR |
 |
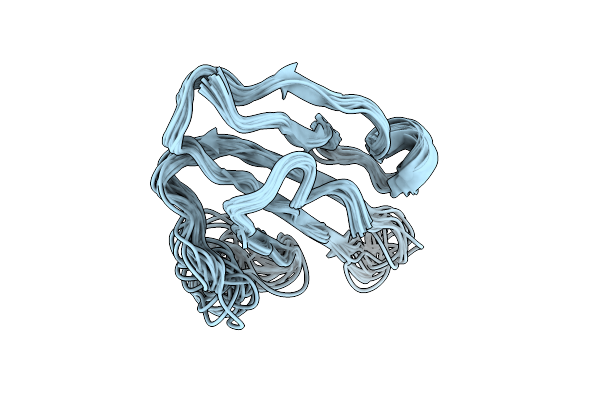
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: POV, CLR |
 |
Solution Structure Of The Human Prostate Stem Cell Antigen (Psca), Water-Soluble Domain
|
 |
Organism: Microbacterium oxydans
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: LIGASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN |

