Search Count: 9,420
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
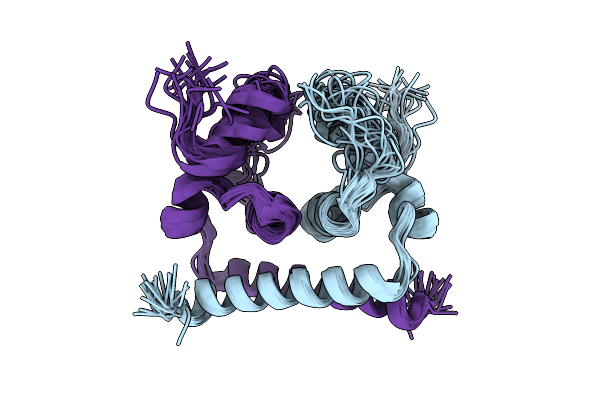 |
Nmr Strucuture Of Dengue Virus 2 Capsid Protein With The Deletion Of The Intrinsically Disordered N-Terminal Region
Organism: Dengue virus type 2
Method: SOLUTION NMR Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
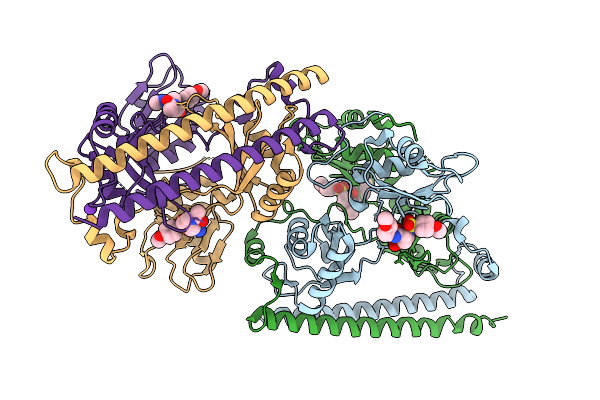 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: A1B7J, K |
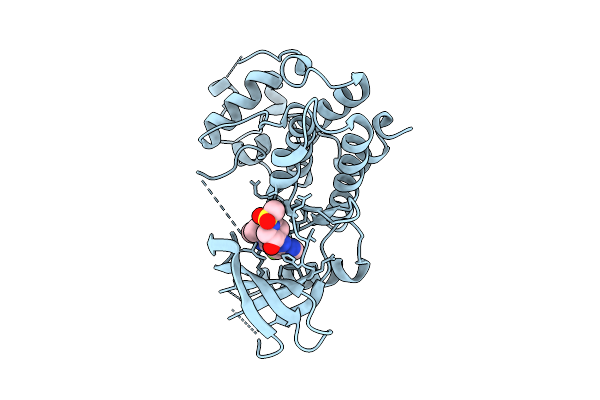 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1CIF |
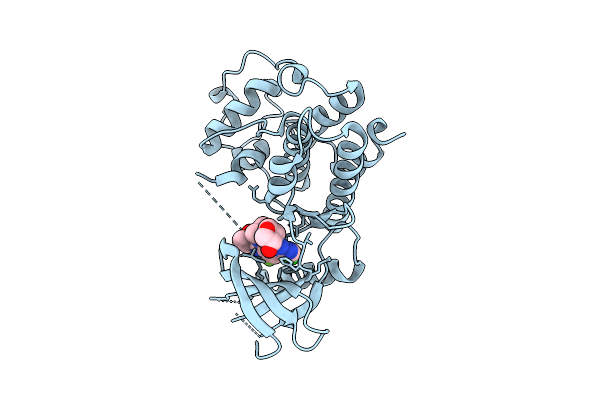 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1AZ4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1CIF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPQ, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPS, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPU |
 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: LIGASE |
 |
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
The Cryo-Em Structure Of Porcine Serum Mgam Bound With Acarviosyl-Maltotriose.
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PEPTIDE NUCLEIC ACID |
 |
Human Impdh2 Mutant - S160Del, Treated With Gtp, Atp, Imp, And Nad+; Tetramer Reconstruction
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: IMP, NAD |
 |
Human Impdh2 Mutant - S160Del, Treated With Gtp, Atp, Imp, And Nad+; Interfacial Octamer Reconstruction
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: IMP, NAD |
 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |

