Search Count: 4,874
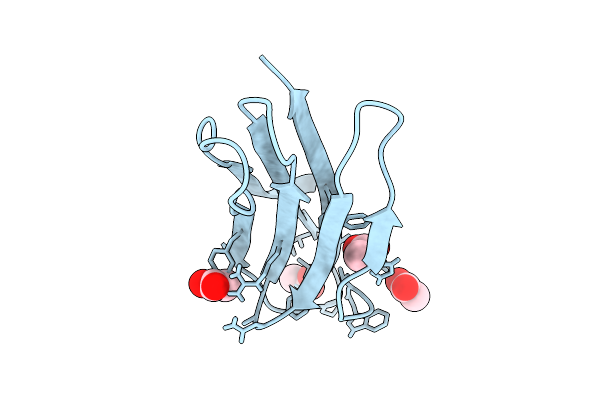 |
Organism: Bacillus licheniformis dsm 13 = atcc 14580
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: ANTIMICROBIAL PROTEIN Ligands: ACT, CL |
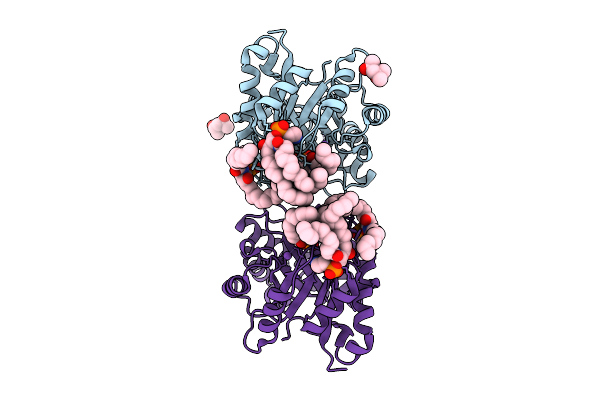 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 2.2 A Resolution From A 2-Day Old Crystal
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
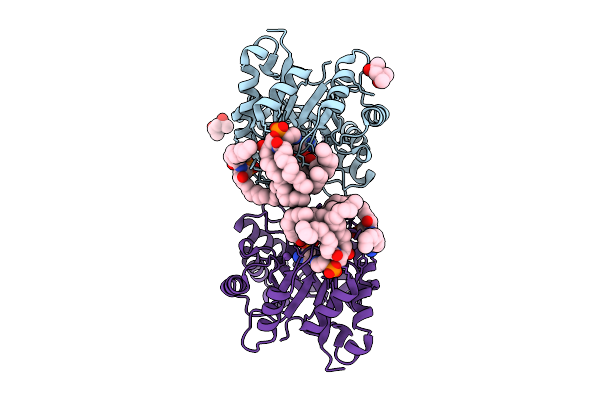 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Substrate Sphingolipids At 2.60 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, NA, MG, MPD |
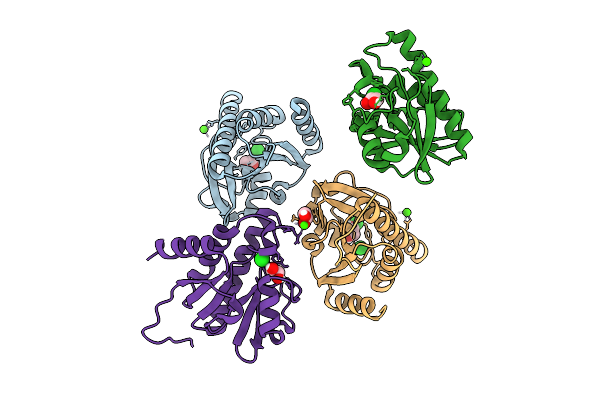 |
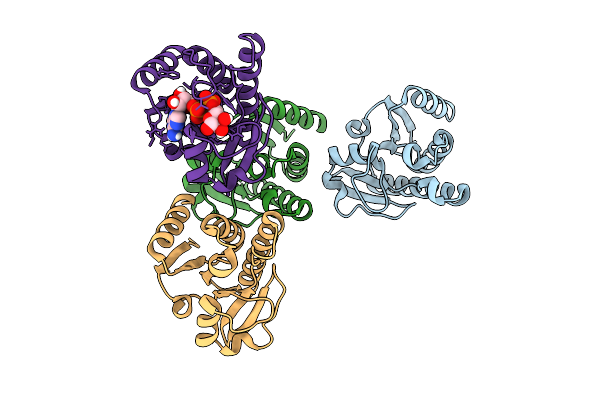
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain D31H Mutant (P31 Crystal Form)
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: EDO, CA, CL |
 |
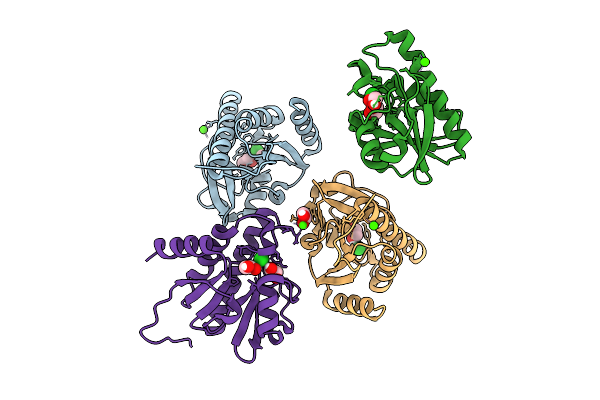
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain D31N Mutant (P41 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: APR |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain D31N Mutant (P31 Crystal Form)
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: EDO, CA, CL |
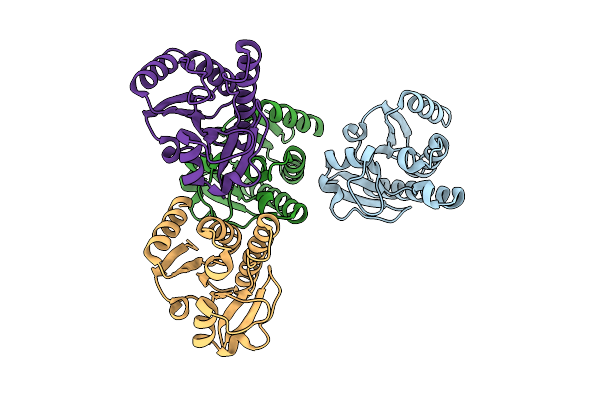 |
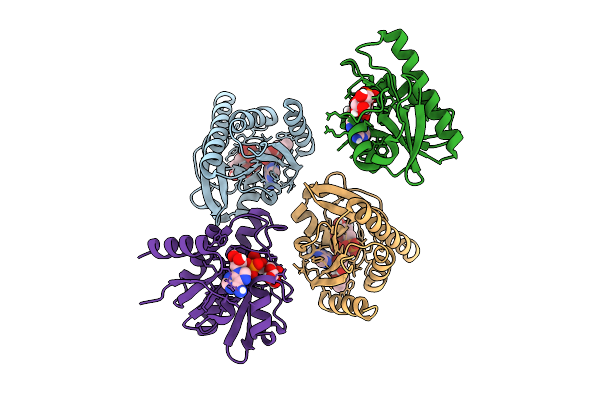
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain D31N Mutant (P41 Crystal Form)
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A Mutant (P31 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: APR, AR6, IPA |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A Mutant (P31 Crystal Form)
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: CL |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A D31H Double Mutant (P31 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: AR6 |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A D31H Double Mutant (P31 Crystal Form)
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A D31N Double Mutant (P31 Crystal Form) In Complex With Adp-Ribose
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: APR, AR6 |
 |
Crystal Structure Of Chikungunya Virus Nsp3 Macrodomain N24A D31N Double Mutant (P31 Crystal Form)
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN,HYDROLASE Ligands: CL |
 |
F420-Dependent Glucose-6-Phosphate Dehydrogenase From Thermomicrobium Roseus With Glucose
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: BGC, NA |
 |
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: CL |
 |
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: SO4, CL |
 |
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: BG6, PEG, PG4, PGE, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: VIRAL PROTEIN,HYDROLASE/INHIBITOR Ligands: A1A55 |
 |
The Cryo-Em Structure Of Human Tissue Nonspecific Alkaline Phosphatase (Htnap) In Complex With Mls-0038949
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE Ligands: MG, ZN, CA, A1JNY |
 |
Organism: Homo sapiens, Synthetic construct, Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MOTOR PROTEIN Ligands: GTP, MG, EDO, GDP, ANP |

