Search Count: 24,054
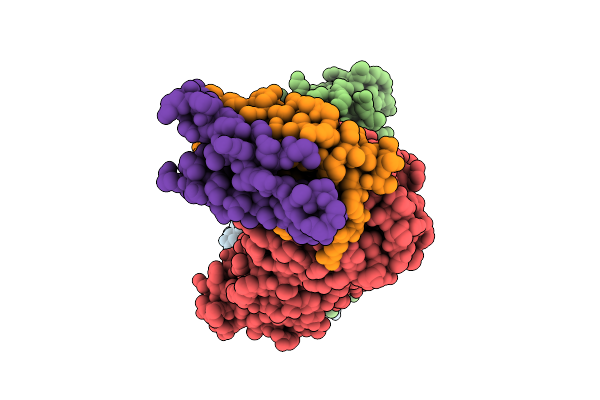 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN |
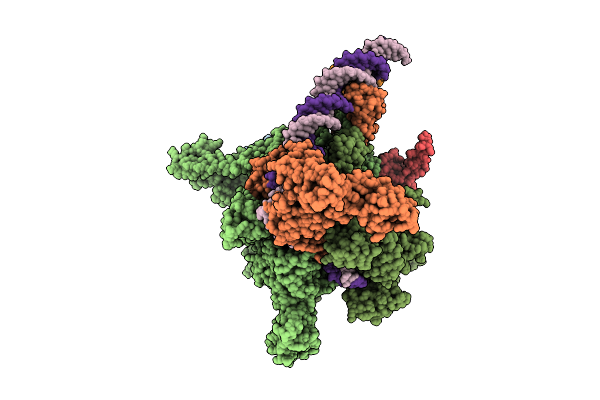 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
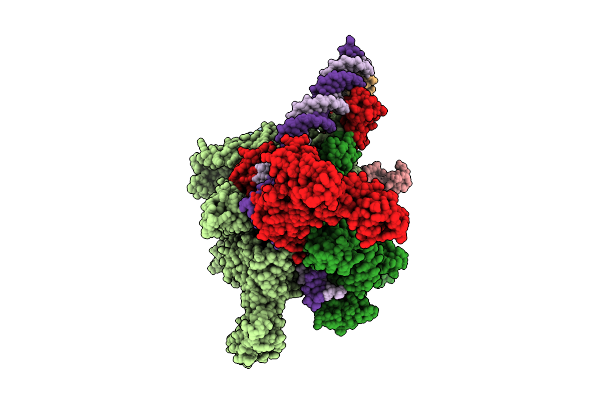 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment And 5-Mer Rna
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
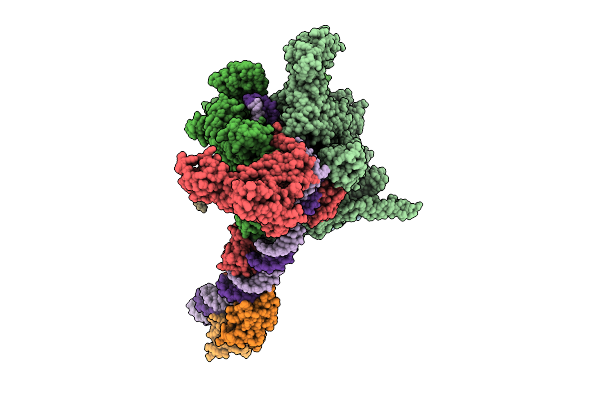 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr Transcription Factor And Ompu Promoter Dna
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
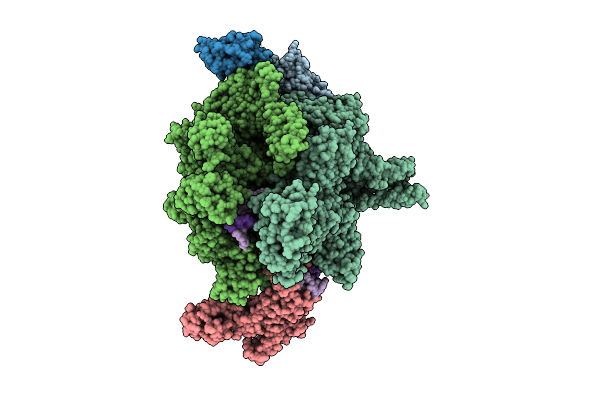
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Tcpp Transcription Factor And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
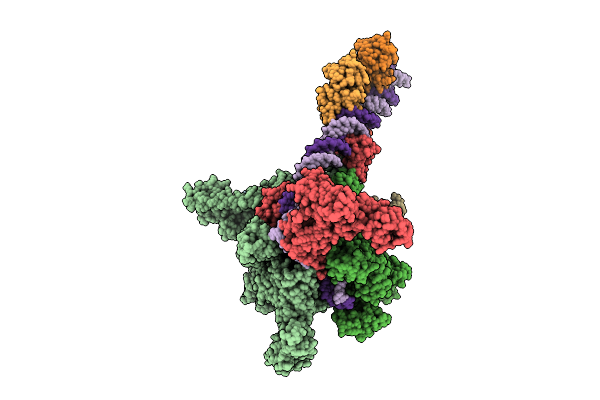
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr And Tcpp Transcription Factors And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
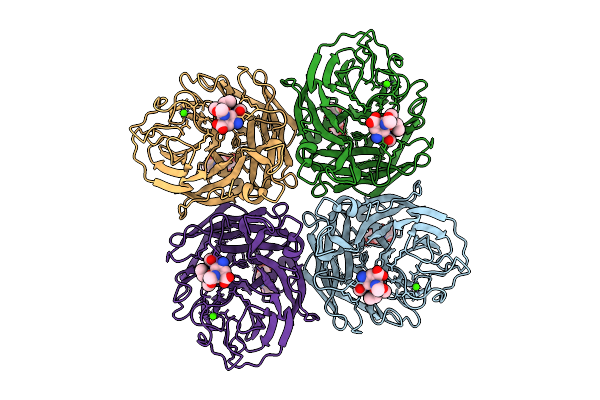 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1IVV, NAG, CA |
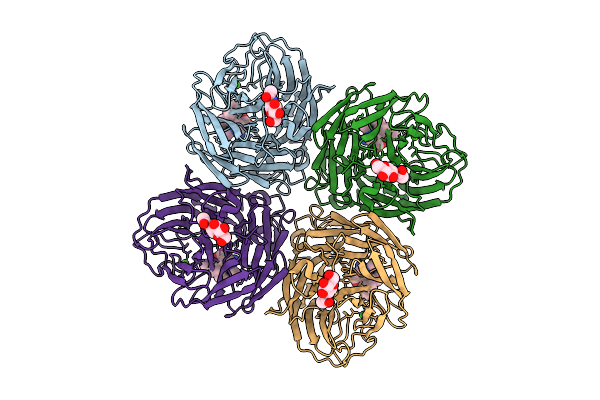 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1IVW, NAG, CA |
 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1IVX, NAG, CA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK3, GOL |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: CO |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPQ, SO4 |

