Search Count: 13,333
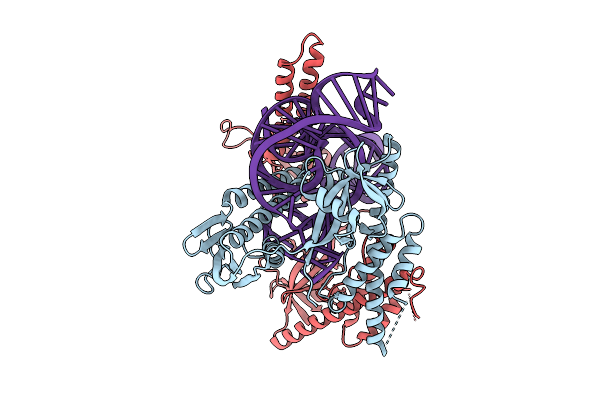 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
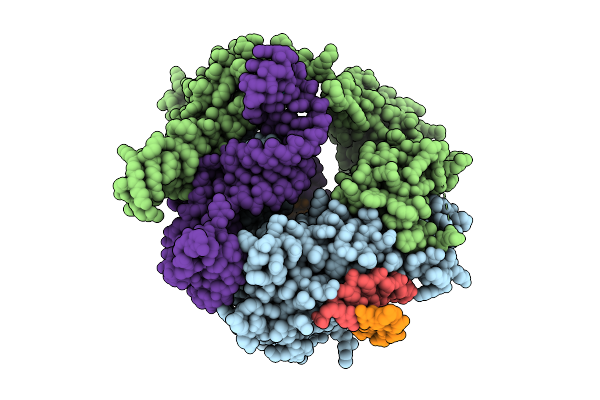 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA |
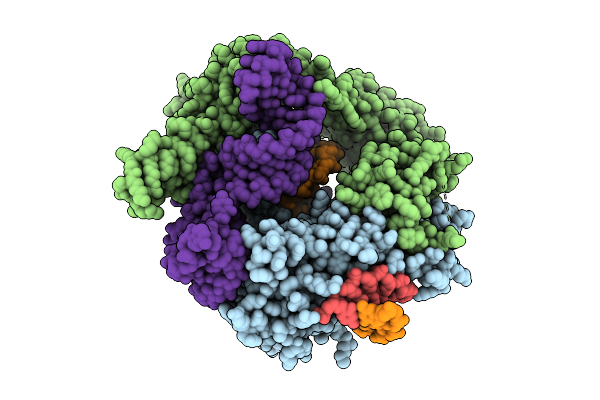 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA |
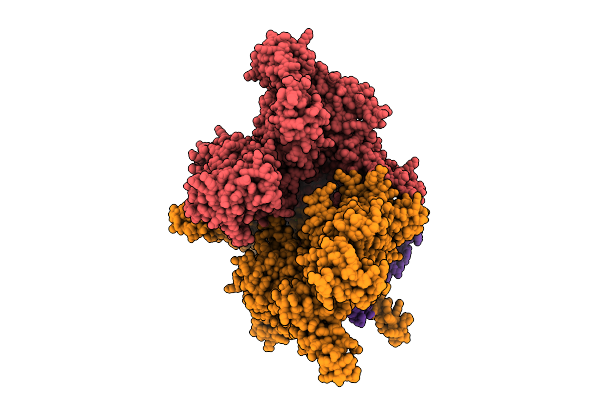 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
 |
Organism: Streptomyces peucetius subsp. caesius atcc 27952
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: HEM, DM1, SO4 |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1JAP, L1P, A1JAO, NA, A1JAN |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: B13 |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: B13, A1JAP, L1P, A1JAO, NA, A1JAN |
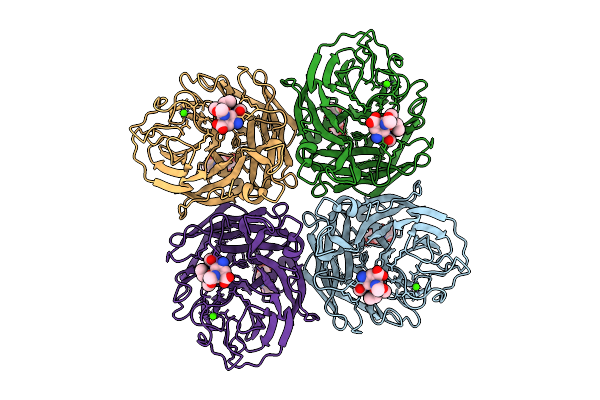 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1IVV, NAG, CA |
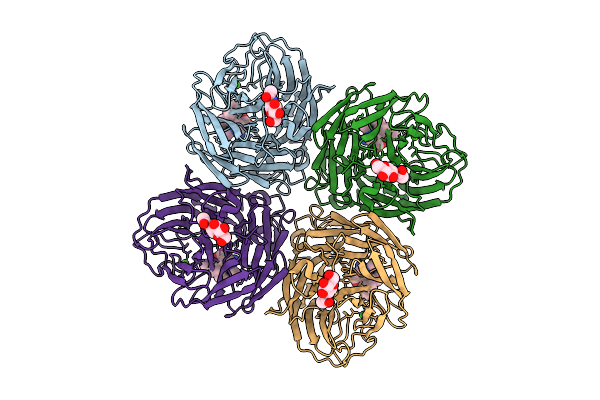 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1IVW, NAG, CA |
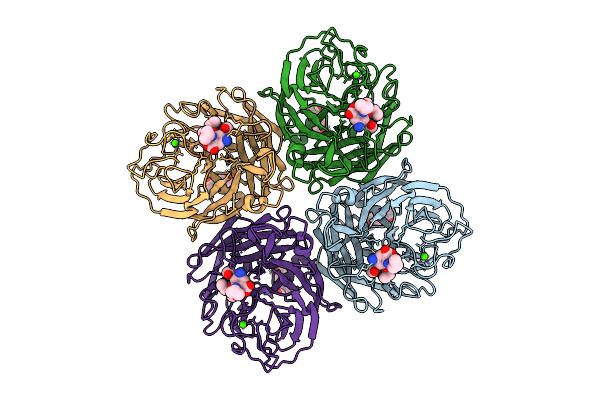 |
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN Ligands: A1IVX, NAG, CA |
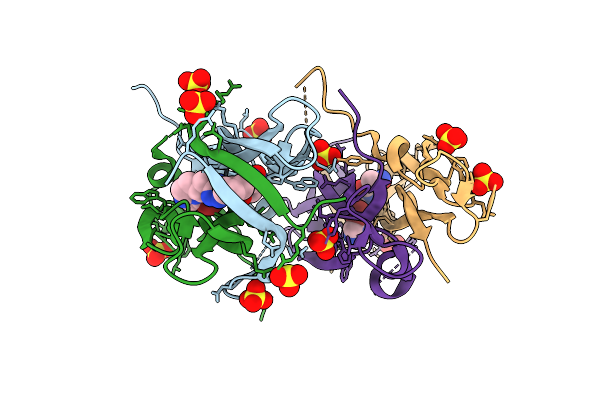 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPQ, SO4 |
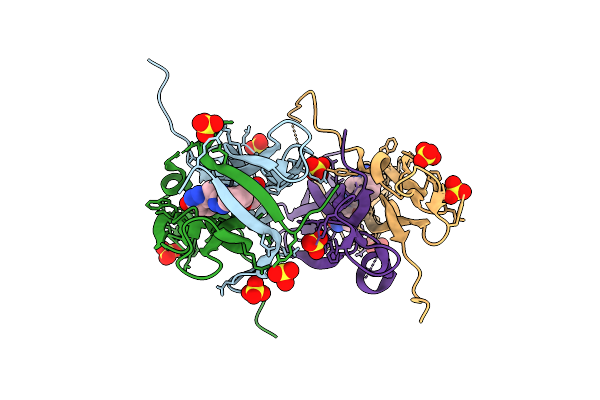 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPS, SO4 |
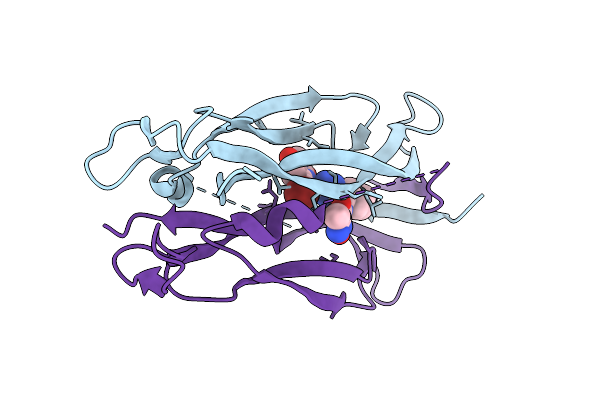 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPU |
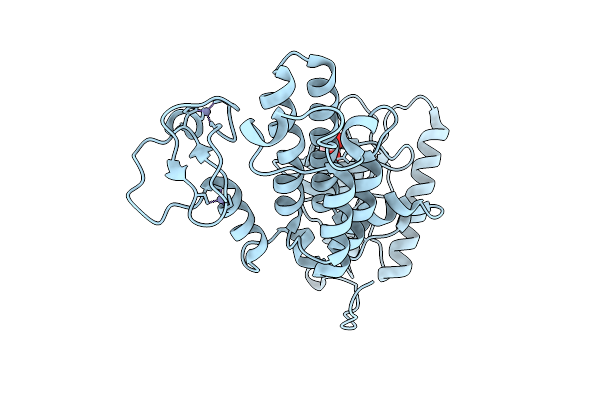 |
Crystal Structure Of Human Prkcbp1 Zinc Finger Mynd-Type Containing 8 With Pentaglutamate Tag
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: EDO, ZN |
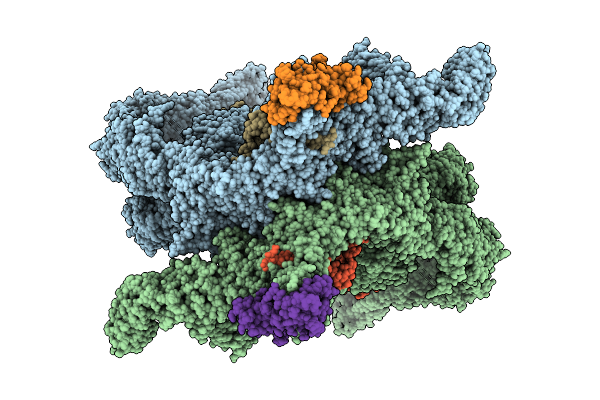 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN |
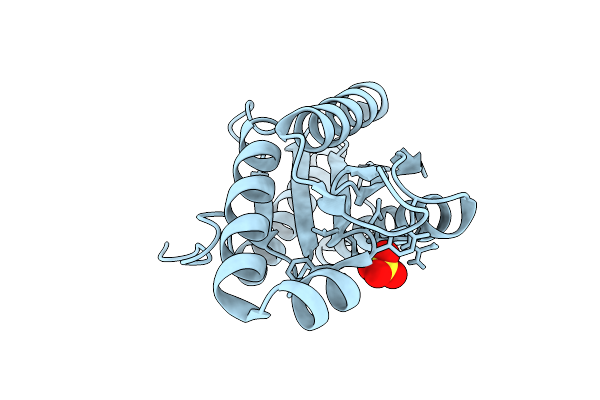 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
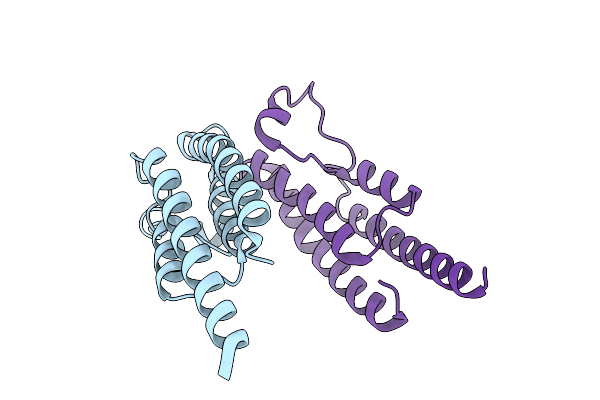 |
Bromodomain Containing Protein 1 With Crystal Epitope Mutations P566E:V569R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: GENE REGULATION |

