Search Count: 22,791
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GDP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GNP |
 |
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: NAG, NA, PEG, EDO, GOL |
 |
Structure Of Lysozyme-N,N',N"-Triacetylchitotriose Complex Determined Using Reyes For Automated Microed
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: CL, NA |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dfph-A.01_10R59P_Lc Fab
Organism: Human immunodeficiency virus 1, Macaca
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
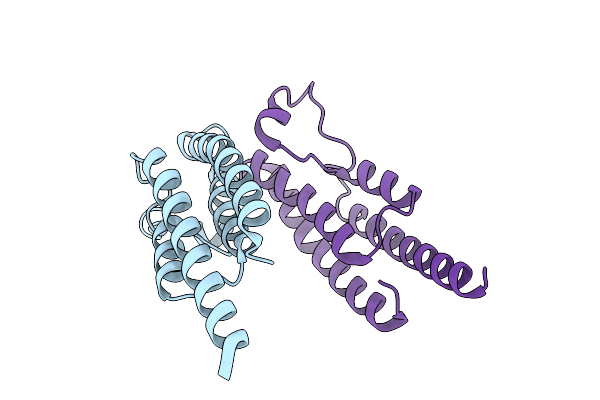
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
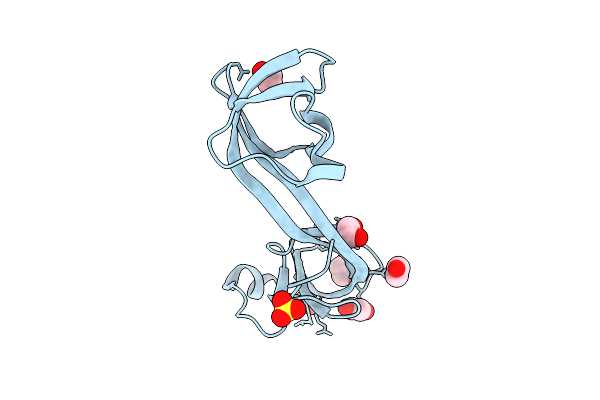
Bromodomain Containing Protein 1 With Crystal Epitope Mutations P566E:V569R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: GENE REGULATION |
 |
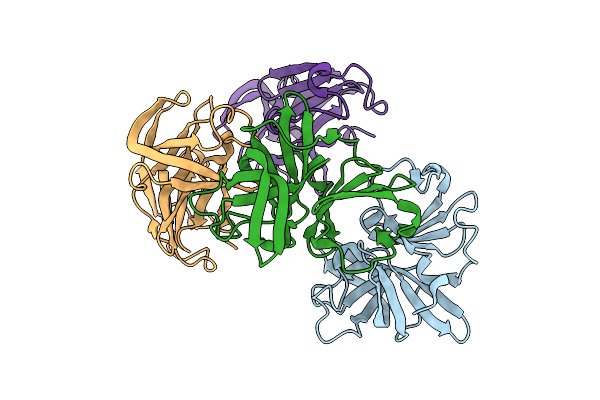
Jumonji Domain-Containing Protein 2B With Crystallization Epitope Mutations L916G:R917A:A918D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: EDO, SO4 |
 |
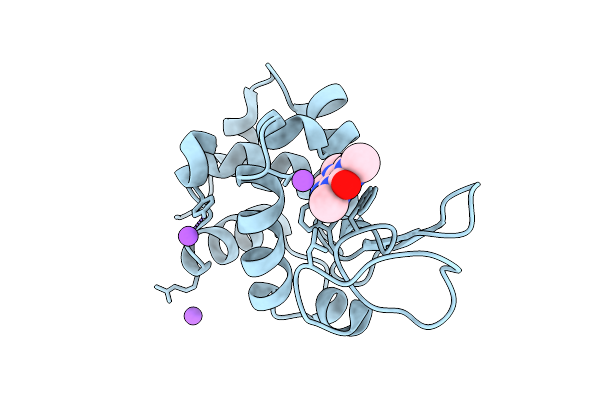
Protease From Norovirus Sydney Gii.4 Strain With Crystallization Epitope Mutation H50Y
Organism: Norovirus sydney 2212
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: SIGNALING PROTEIN |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: SIGNALING PROTEIN Ligands: MG |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: SIGNALING PROTEIN |
 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-12-03 Classification: DE NOVO PROTEIN |
 |
Backbone-Modified Parallel Beta Hairpin (Pbh): N-Alpha-Amino Arginine At Position 17
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-12-03 Classification: DE NOVO PROTEIN |

