Search Count: 7,192
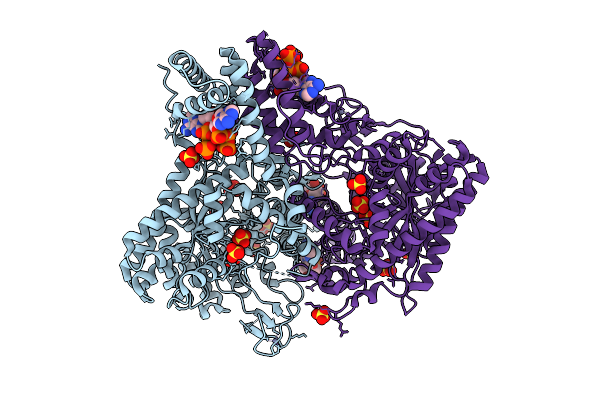 |
Crystal Structure Of S. Thermophilus Class Iii Ribonucleotide Reductase Bound To Datp
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: MG, DTP, ZN, SO4 |
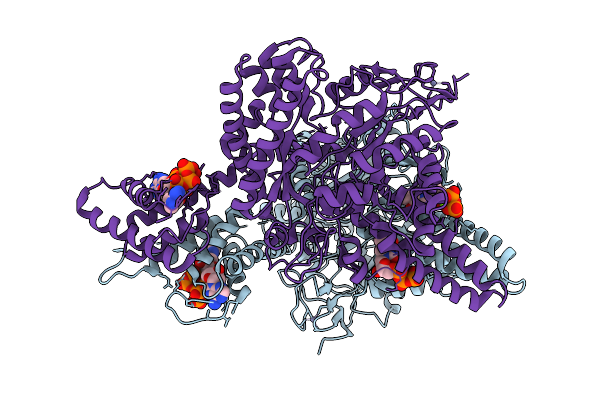 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: TTP, DTP, MG, ZN |
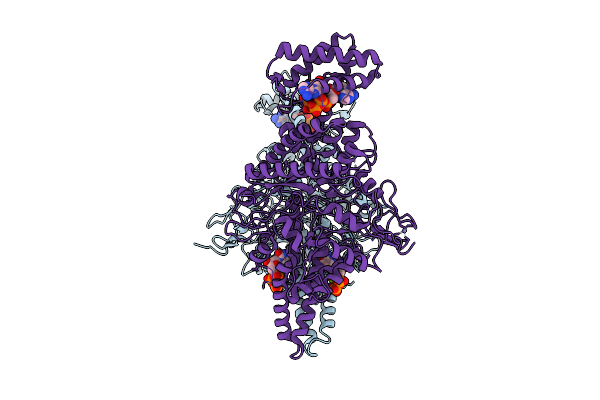 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: ATP, MG, TTP, ZN |
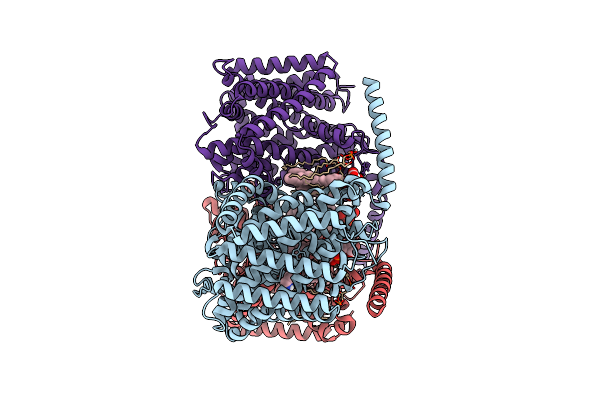 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: ABU, PGT, CDL |
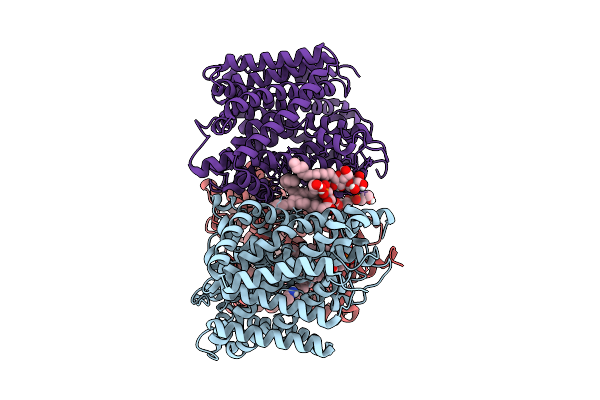 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: PGT, ABU, LMT |
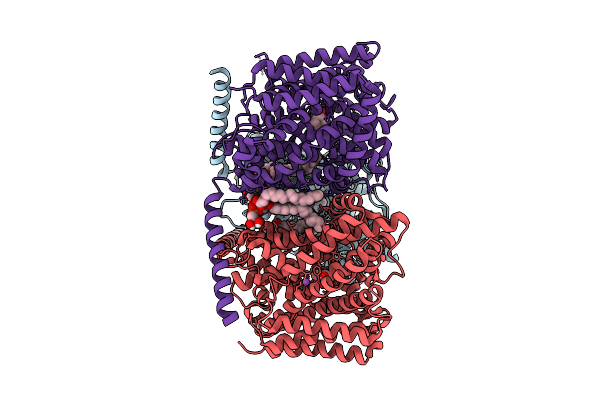 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: PGT, BET, NA |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
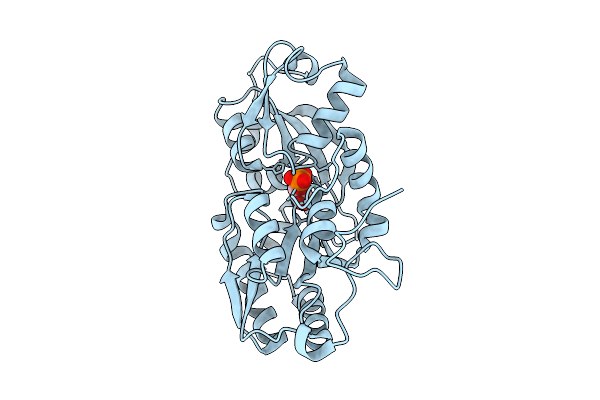
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN/DNA |
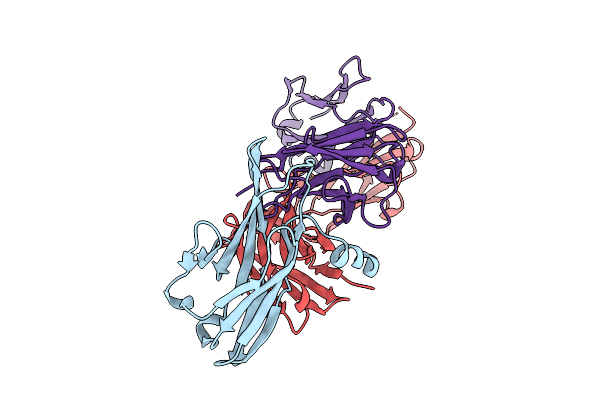 |
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
 |
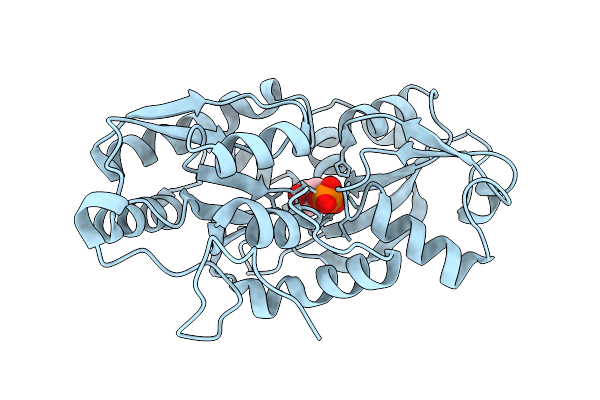
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
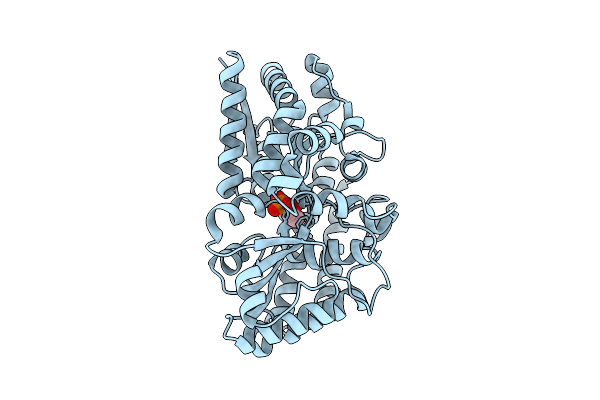
Organism: Marinobacter sp. dsm 11874
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
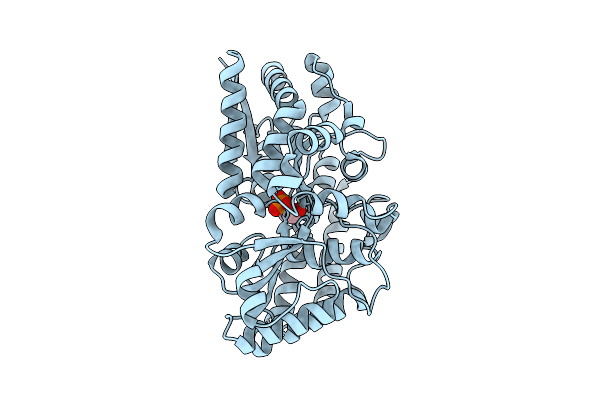
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: 1GP |
 |
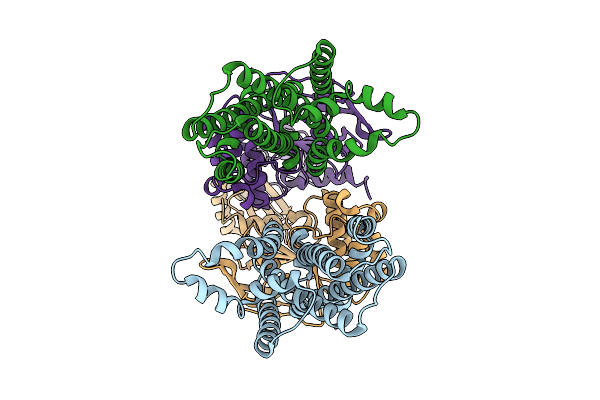
Organism: Phaeobacter sp. med193
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: G3P |
 |
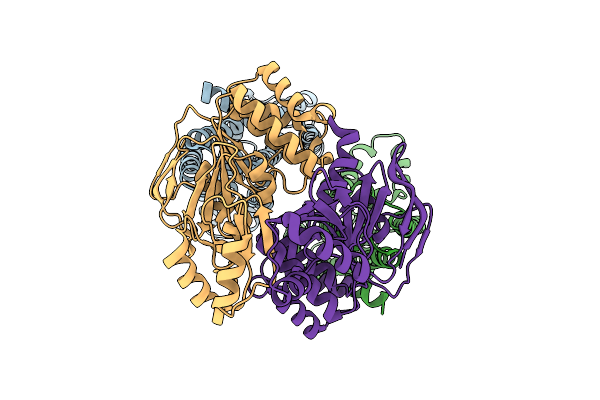
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
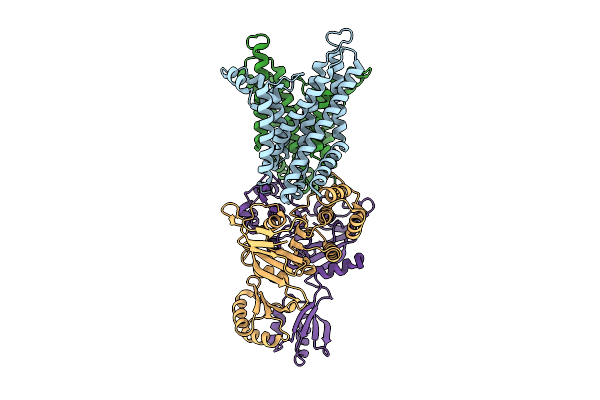 |
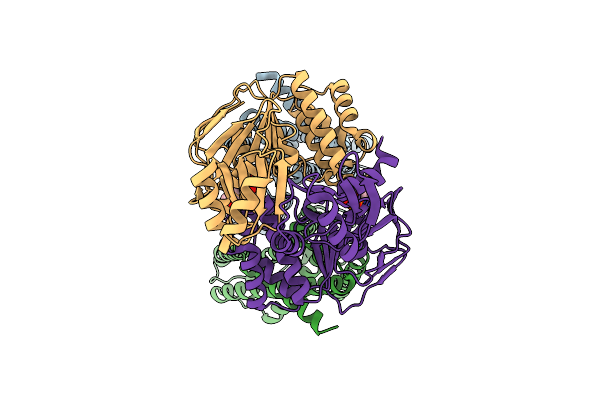
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
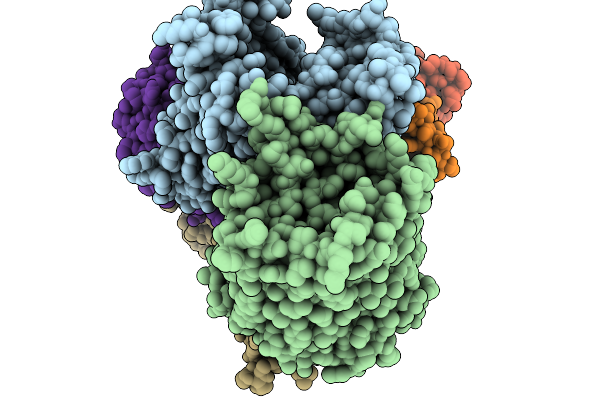
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: PLM, Z41 |

