Search Count: 3,113
 |
Structure Of Cathepsin B1 From Schistosoma Mansoni (Smcb1) In Complex With A Carborane Inhibitor
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: A1IHQ, EDO |
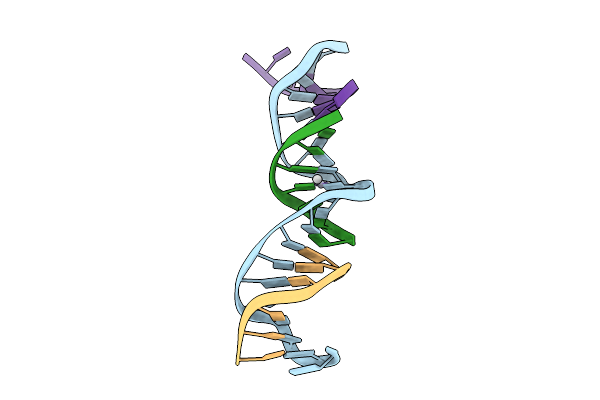 |
[T:Ag+/Hg2+:T--(Ph9.5-Ph8; 4S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 9.5 And Soaked In Ph 8 For 4S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: HG, AG |
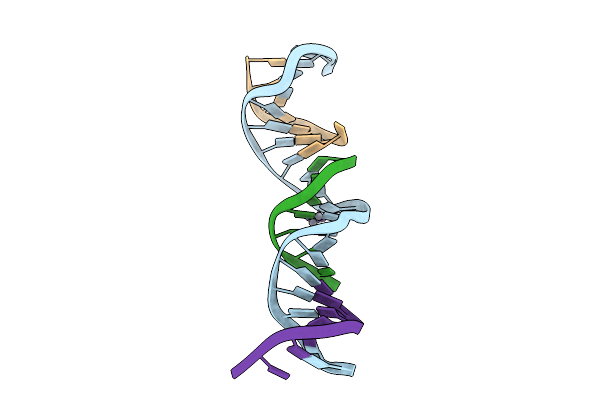 |
[T:Ag+/Hg2+:T--(Ph9.5-Ph8; 7S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 9.5 And Soaked In Ph 8 For 7S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: HG |
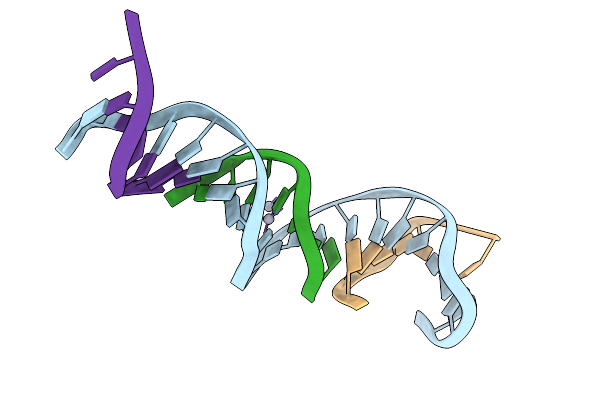 |
[T:Ag+/Hg2+:T--(Ph8-Ph9.5; 4S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 8 And Soaked In Ph 9.5 For 4S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: HG |
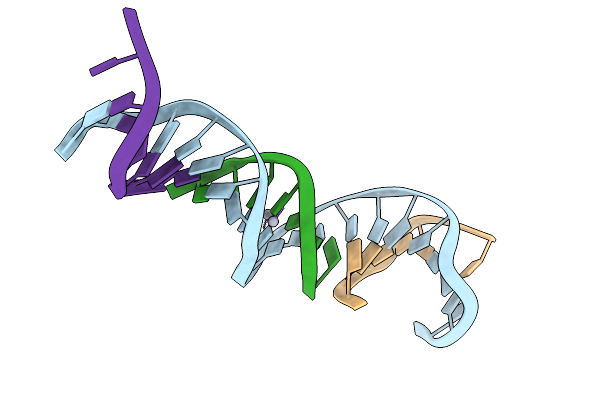 |
[T:Ag+/Hg2+:T--(Ph8-Ph9.5; 16S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 8 And Soaked In Ph 9.5 For 16S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: HG |
 |
[T:Ag+/Hg2+:T--(Ph8-Ph9.5; 35S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 8 And Soaked In Ph 9.5 For 35S
Organism: Synthetic construct, Synthetic construct (code 12)
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: HG, AG |
 |
[T:Ag+/Hg2+:T--(Ph9.5-Ph11; 20S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 9.5 And Soaked In Ph 11 For 20S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: AG |
 |
[T:Ag+/Hg2+:T--(Ph9.5-Ph11; 40S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 9.5 And Soaked In Ph 11 For 40S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: AG |
 |
[T:Ag+/Hg2+:T--(Ph9.5-Ph11; 7S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 9.5 And Soaked In Ph 11 With Ag+ And Hg2+ For 7S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: AG |
 |
[T:Ag+/Hg2+:T--(Ph9.5-Ph11; 21S)] Metal-Mediated Dna Base Pair In Tensegrity Triangle Grown At Ph 9.5 And Soaked In Ph 11 With Ag+ And Hg2+ For 21S
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DNA Ligands: AG |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: EDO, MES |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: MG, CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: CL |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: SO4, ACT, GOL, CL, NA |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: GOL |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CL, GOL, ACT |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CL, A1EGH, ACT, PO4 |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: A1EGL, CL |
 |
Organism: Leptinotarsa decemlineata
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: MG, EDO, A1JD4 |

