Search Count: 3,811
 |
Organism: Priestia megaterium, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
The Cryo-Em Structure Of Porcine Serum Mgam Bound With Acarviosyl-Maltotriose.
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Protease From Norovirus Sydney Gii.4 Strain With Crystallization Epitope Mutation H50Y
Organism: Norovirus sydney 2212
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN |
 |
Organism: Rift valley fever virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: GOL, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL ADHESION Ligands: GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL ADHESION Ligands: GOL, FMT, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL ADHESION Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL CYCLE Ligands: EDO |
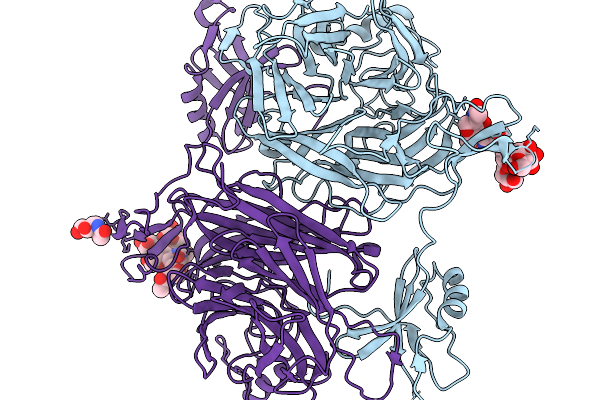 |
Organism: Jeilongvirus beilongi
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: VIRAL PROTEIN Ligands: NAG, BMA, MAN |
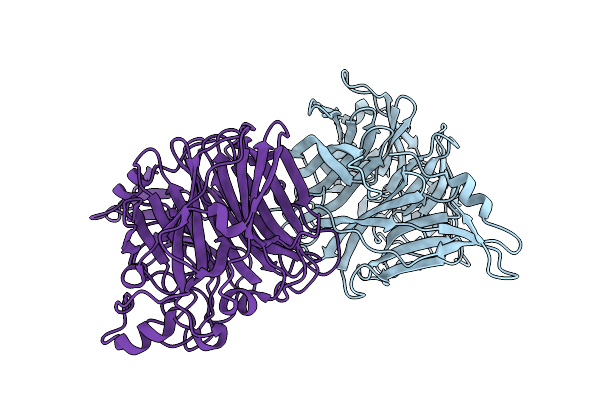 |
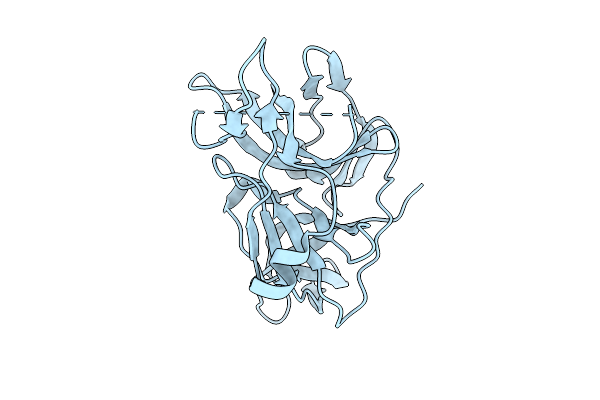 |
Dengue 2 Virus Ns2B-Ns3 Protease Fusion Protein With Crystal Epitope Mutation K174Q
Organism: Dengue virus type 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
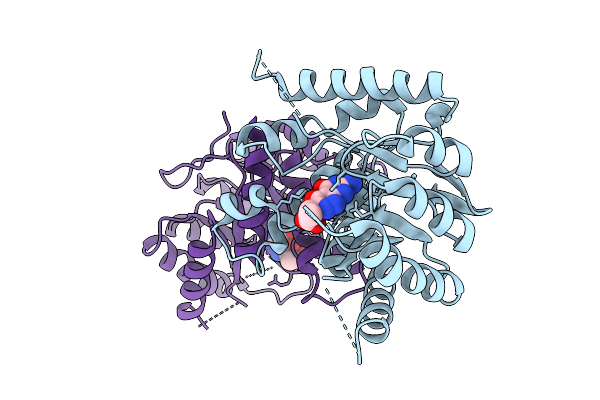 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: A1CG3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: A1CG7 |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: ZN |

