Search Count: 18,409
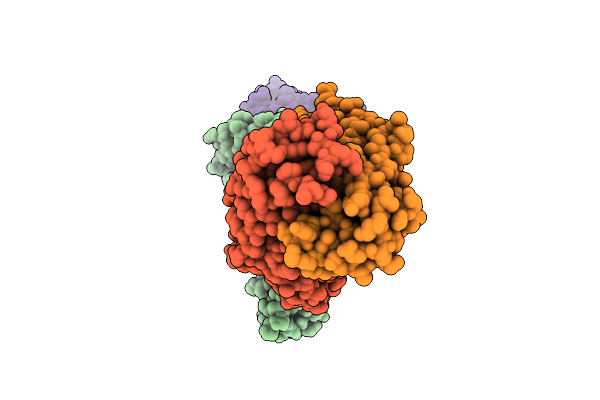 |
Organism: Mus musculus, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: PROTEIN BINDING |
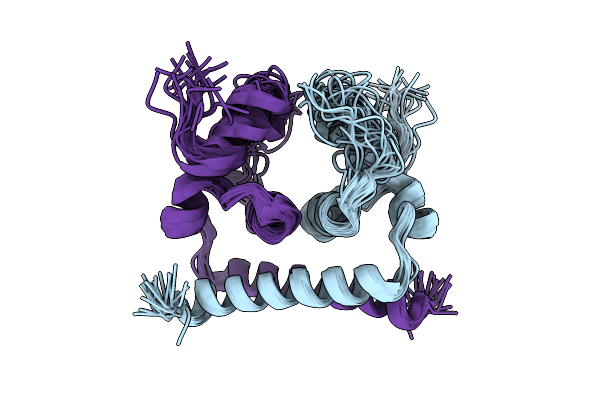 |
Nmr Strucuture Of Dengue Virus 2 Capsid Protein With The Deletion Of The Intrinsically Disordered N-Terminal Region
Organism: Dengue virus type 2
Method: SOLUTION NMR Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
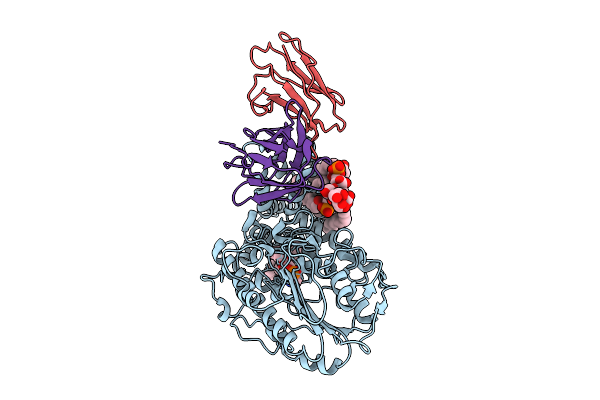 |
Single-Particle Cryo-Em Structure Of The First Variant Of Mobilized Colistin Resistance (Mcr-1) In Its Ligand-Bound State
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: KDL, PEE |
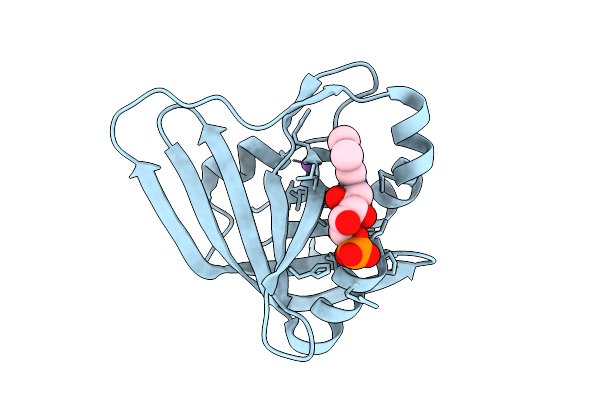 |
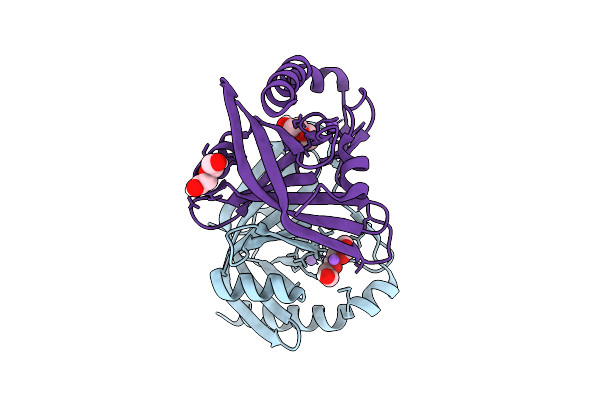
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: DNA Ligands: LDP |
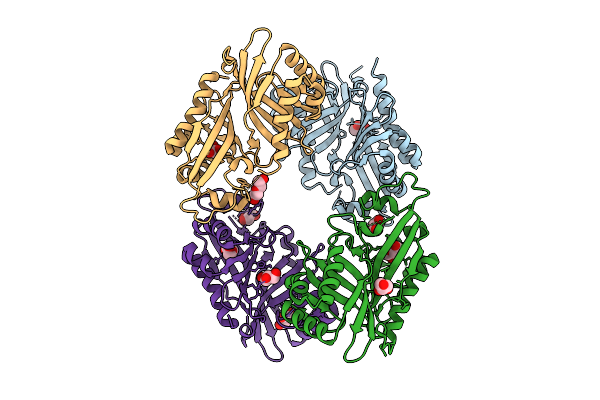 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
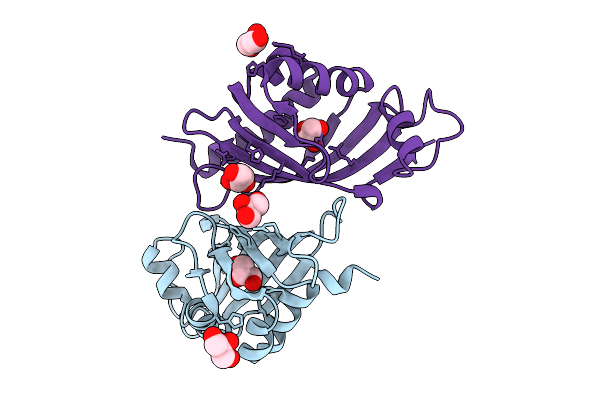 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
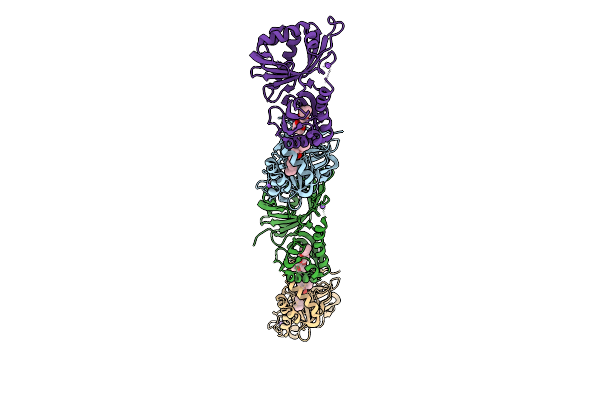 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
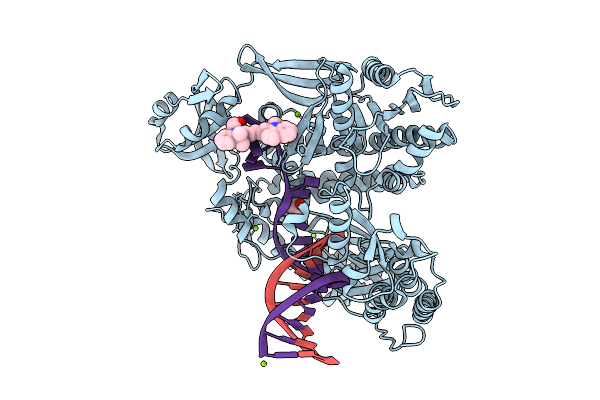 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG, 5CY, GOL |
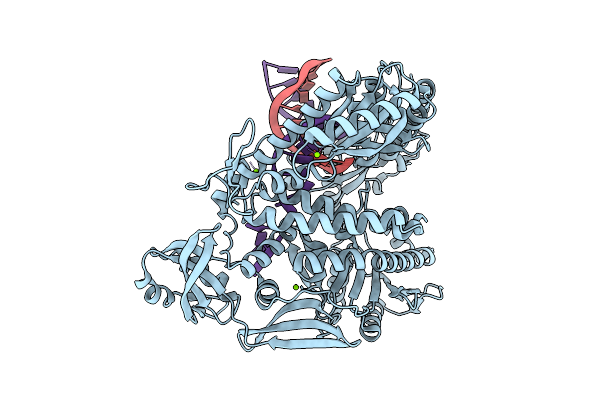 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
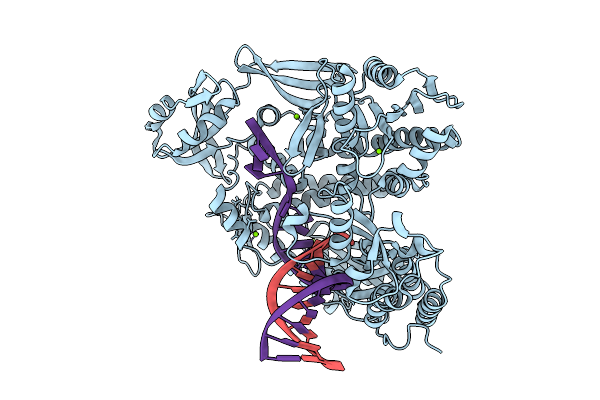 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
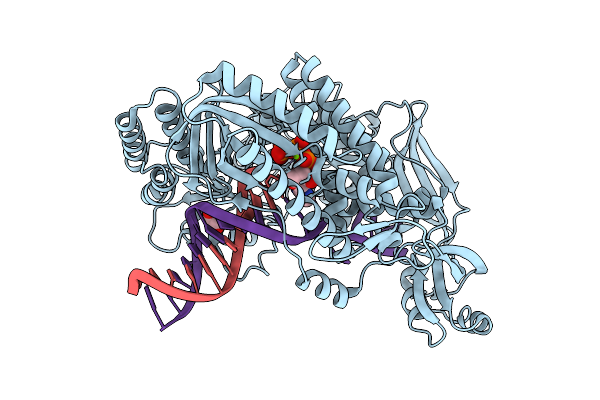 |
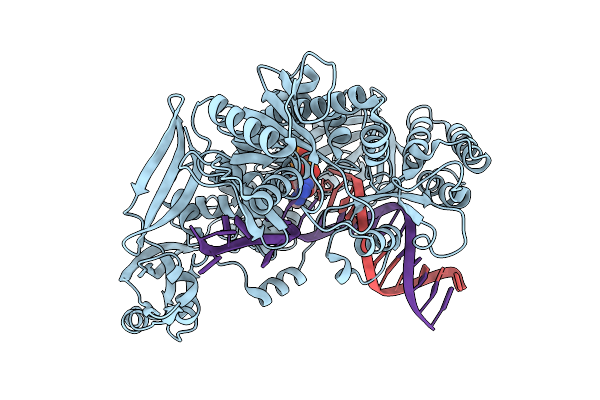
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, EDO |
 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, CL |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ADP, MG |
 |
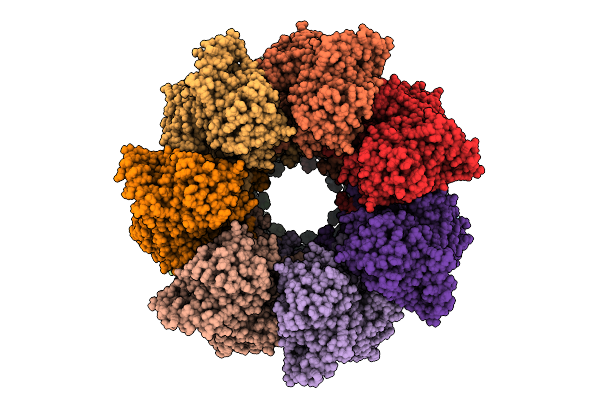
Ancestral Group Iii Chaperonin (Aciii) Double-Ring In Open Conformation Including The Equatorial And Intermediate Domains (Residues 12-200 And 356-507)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Ancestral Fibrobacteres-Chlorobi-Bacteroidetes Group Chaperonin (Afcb) Double-Ring In Open Conformation
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE |
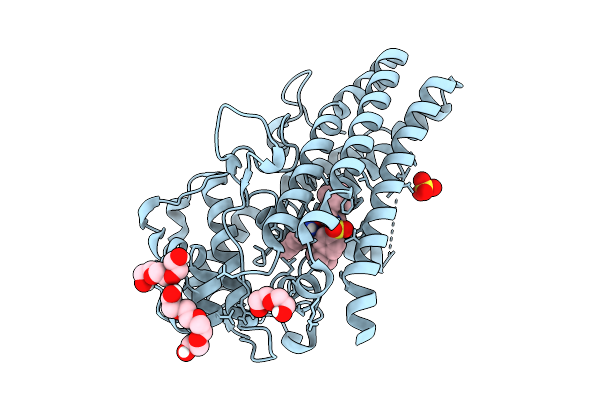 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: A1JLE, SO4, PG4, PGE, PEG |
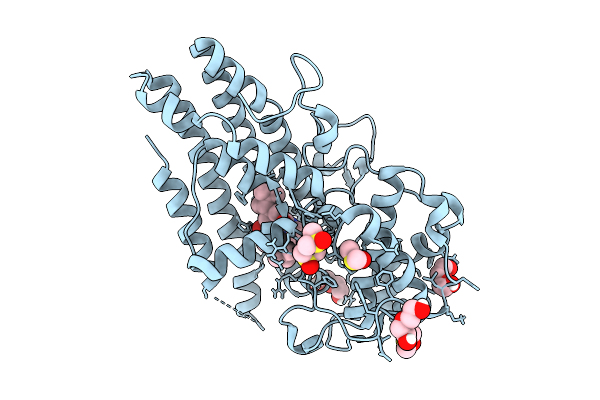 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: PEG, A1JLD, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: A1JLB, PEG |

