Search Count: 79,531
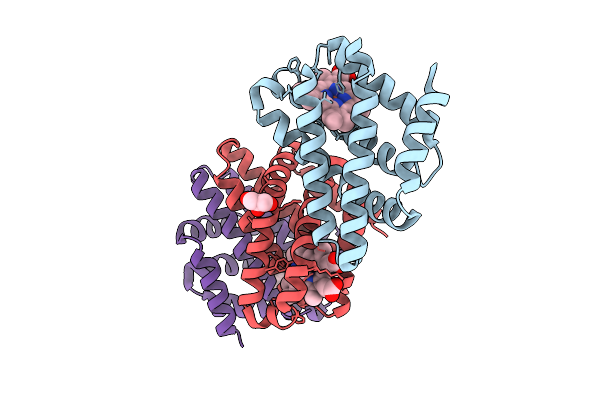 |
Cyanide-Ligated Bordetella Pertussis Globin Coupled Sensor Regulatory Domain
Organism: Tohama i / atcc baa-589 / nctc 13251
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SIGNALING PROTEIN Ligands: HEM, CYN, GOL |
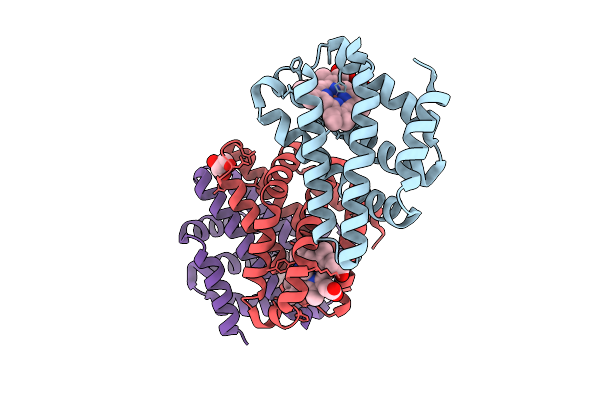 |
Cyanide-Ligated Bordetella Pertussis Globin Coupled Sensor Regulatory Domain S68A
Organism: Bordetella pertussis (strain tohama i / atcc baa-589 / nctc 13251)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SIGNALING PROTEIN Ligands: HEM, CYN, GOL |
 |
Three-Dimensional Structure Of The Merozoite Surface Protein 1 C-Terminal Domain
Organism: Plasmodium berghei
Method: SOLUTION NMR Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: A2G |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: GOL, A2G |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN |
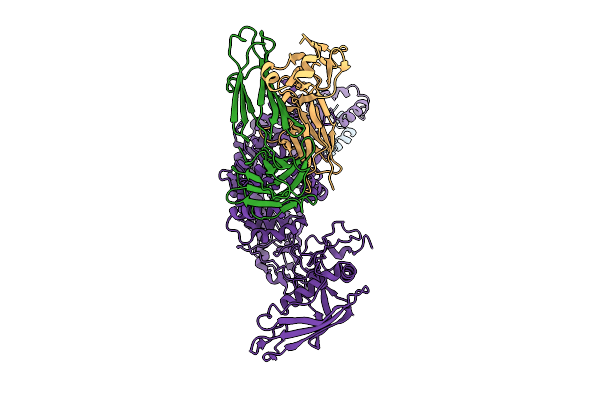 |
P116 From Mycoplasma Pneumoniae In Complex With Mild Growth Suppressor Monoclonal Antibody
Organism: Mus musculus, Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of The Human Frataxin Protein In Complex With A Tailored Camelid Nanobody 6B1
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE |
 |
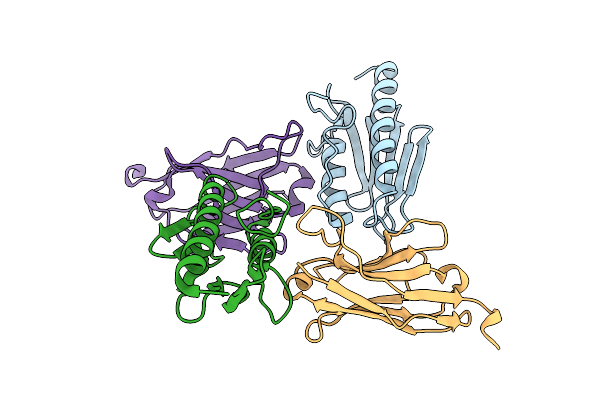
Crystal Structure Of The Human Frataxin Protein In Complex With A Tailored Camelid Nanobody 4A7
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The Human Frataxin Protein In Complex With A Tailored Camelid Nanobody 16C10
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE |
 |
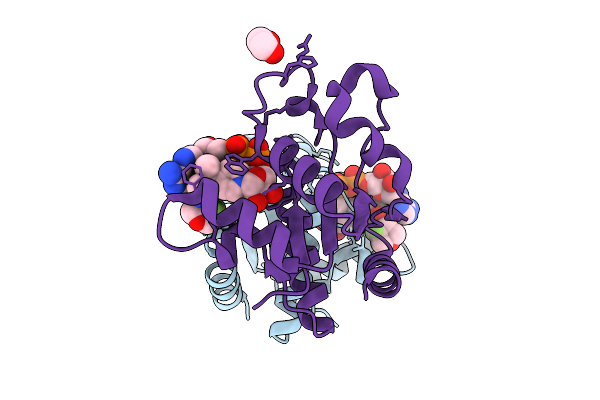
[Fefe]-Hydrogenase From D. Desulfuricans With Synthetic Active Site Containing Only One Cyanide Ligand.
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: SF4, A1IW2, CL, LI |
 |
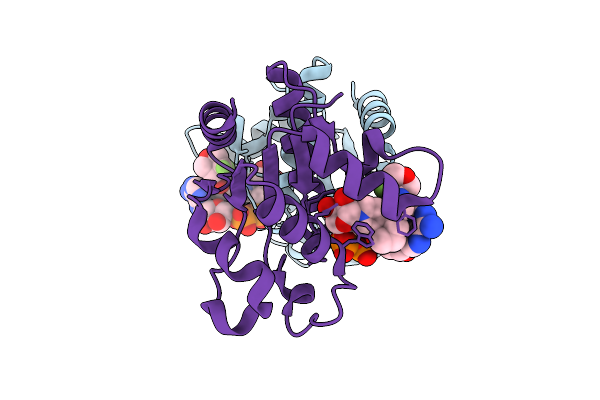
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1IW3, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1IWL |
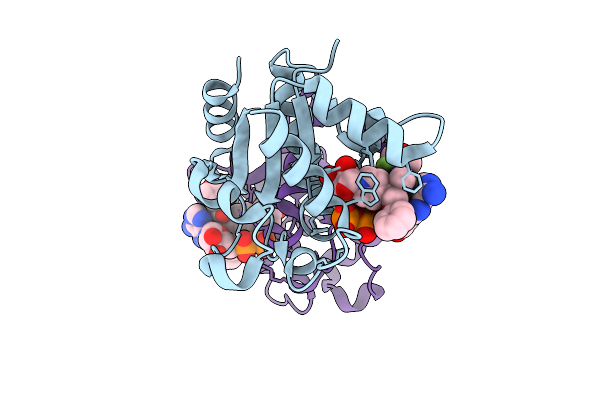 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1IWN |
 |
Crystal Structure Of An Allosteric Inhibitor Bound To Human Ripk1 Kinase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1IX7, IOD, 1HX |
 |
Crystal Structure Of An Allosteric Inhibitor Bound To Human Ripk1 Kinase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1IX9, IOD, PEG |
 |
Crystal Structure Of Brd2 Bd2 Domain In Complex With Small Molecule Inhibitor Mivebresib Abbv-075
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: 8NG |
 |
Crystal Structure Of Brd2 Bd1 Domain In Complex With Small Molecule Inhibitor Isoxazole Azepine Compound.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: 1XB |
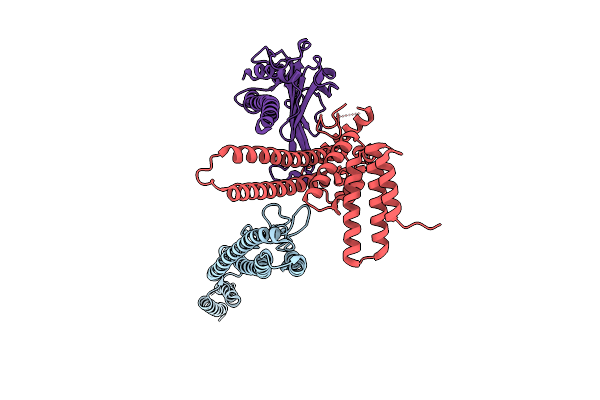 |
Straight And Symmetrical Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
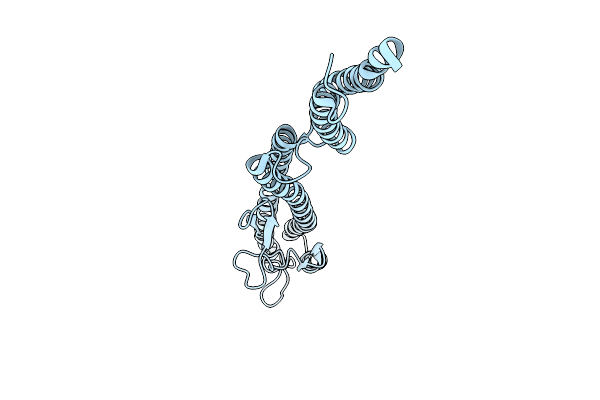 |
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |

