Search Count: 216
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2025-04-23 Classification: PLANT PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2025-04-23 Classification: PLANT PROTEIN |
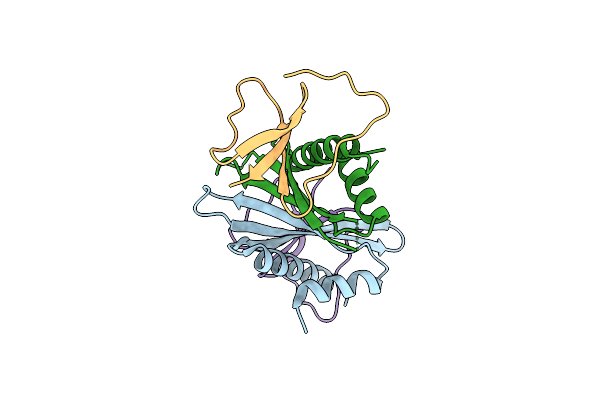 |
The Crystal Structure Of The Minimal Interaction Domains Of Drb7.2:Drb4 Complex
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2024-06-26 Classification: GENE REGULATION |
 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: RNA BINDING PROTEIN |
 |
Rauvolfia Serpentina Strictosidine Synthase (Rsstr) In Complex With A Non-Reactive Tryptamine Substitute Crystallized In P1211 Space Group
Organism: Rauvolfia serpentina
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-02-28 Classification: LYASE Ligands: F66, PEG |
 |
Crystal Structure Of Thiomorpholine-Carboxylate Dehydrogenase From Candida Parapsilosis.
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: EDO, NA, CL, MG |
 |
Crystal Structure Of A Variable Region Segment Of Leptospira Host-Interacting Outer Surface Protein, Liga
Organism: Leptospira interrogans
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-06-28 Classification: CELL ADHESION Ligands: K, CL, EDO, PO4, NA, CA, IOD |
 |
Crystal Structure Of The N-Terminal Domain Of Mutants Of Human Apolipoprotein-E (Apoe)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-20 Classification: LIPID TRANSPORT |
 |
Crystal Structure Of The N-Terminal Domain Of Mutants Of Human Apolipoprotein-E (Apoe)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: LIPID TRANSPORT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2021-08-25 Classification: LIGASE Ligands: ZN |
 |
Organism: Frankia sp. cc1.17
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-08-18 Classification: ISOMERASE Ligands: EDO, DMS |
 |
Organism: Streptomyces rugosporus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-08-18 Classification: ISOMERASE Ligands: HOF, GOL |
 |
Crystal Structure Of Human Methionine Aminopeptidase (Hsmetap1B) In Complex With An-P2-5H-06
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2021-02-24 Classification: HYDROLASE Ligands: CO, EYF, GOL, NA, MG |
 |
Crystal Structure Of Human Methionine Aminopeptidase (Hsmetap1B) In Complex With Kv-P2-4H-05
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-02-24 Classification: HYDROLASE Ligands: CO, EYL, NA, GOL, MG |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-02-24 Classification: HYDROLASE Ligands: CO |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-01-13 Classification: DNA BINDING PROTEIN/DNA |

