Search Count: 192
 |
Structure Of Thermotoga Maritima Gh5 Endoglucanase Tm1752 In Complex With Tris
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2022-03-30 Classification: HYDROLASE Ligands: TRS, IPA |
 |
Structure Of The Thermotoga Maritima Family 5 Endo-Glucanase In Complex With 1-Deoxynojiromycin
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: NOJ, IPA |
 |
Crystal Structure Of Apo Form Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-09-01 Classification: HYDROLASE |
 |
Crystal Structure Of Nadh Bound Holo Form Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima At 1.97 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI |
 |
Crystal Structure Of Thermotoga Maritima Alpha-Glucuronidase (Tm0752) In Complex With Nadh And D-Glucuronic Acid
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI, BDP, IPA |
 |
Crystal Structure Of Salmonella Typhi Outer Membrane Phospholipase (Ompla) Dimer With Bound Calcium
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: BOG, CA, D12 |
 |
Structure Of Deletion Mutant Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: MN, NAI |
 |
Structure Of Nadh Complex Of Thermotoga Maritima Alpha-Glucuronidase At 2.15 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: NAI |
 |
Structure Of Intact Chitinase With Hevein Domain From The Plant Simarouba Glauca, Known For Its Traditional Anti-Inflammatory Efficacy
Organism: Simarouba glauca
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2020-12-23 Classification: HYDROLASE Ligands: CL, MG |
 |
Structure Of A K245A Mutant Of A Group Ii Plp Dependent Decarboxylase From Methanocaldococcus Jannaschii, In Complex With Plp
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-12-02 Classification: LYASE Ligands: PLP, NH4, GOL, SO4 |
 |
Structure Of A K245A Mutant Of L-Tyrosine Decarboxylase From Methanocaldococcus Jannaschii Complexed With L-Tyr: External Aldimine Form
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: LYASE Ligands: SO4, GOL, 0PR |
 |
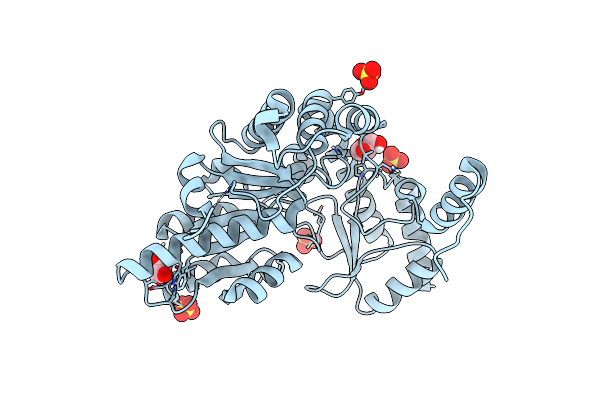
K245A Mutant Of L-Tyrosine Decarboxylase From Methanocaldococcus Jannaschii Complexed With A Post-Decarboxylation Quinonoid-Like Intermediate Formed With L-Tyrosine
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-12-02 Classification: LYASE Ligands: GOL, SO4, EBR |
 |
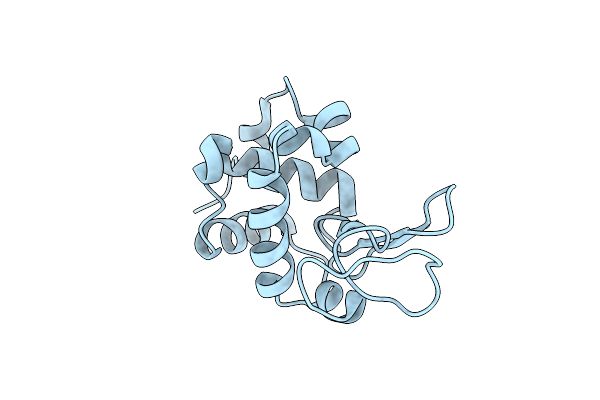
Structure Of A R371A Mutant Of A Group Ii Plp Dependent Decarboxylase From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: LYASE Ligands: SO4, GOL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-10-07 Classification: HYDROLASE Ligands: CL |
 |
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-12 Classification: SUGAR BINDING PROTEIN Ligands: CA, MLI |
 |
Crystal Structure Of Lectin From Pleurotus Ostreatus In Complex With Glycerol
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-08-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
Crystal Structure Of Lectin From Pleurotus Ostreatus In Complex With Rhamnose
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, RAM |
 |
Crystal Structure Of Lectin From Pleurotus Ostreatus In Complex With Galnac
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-08-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL, NGA |
 |
Crystal Structure Of Lectin From Pleurotus Ostreatus In Complex With Galactose
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-08-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL, GLA |
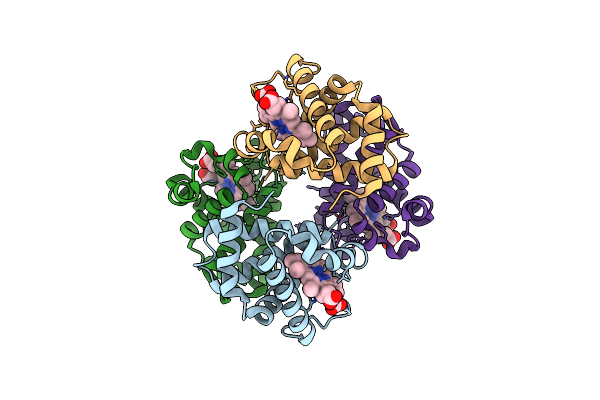 |
Crystal Structure Of Hemoglobin From Turkey (Meleagiris Gallopova) Crystallized In Orthorhombic Form At 1.4 Angstrom Resolution
Organism: Meleagris gallopavo
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-07-22 Classification: OXYGEN STORAGE/OXYGEN TRANSPORT Ligands: HEM, NA |

