Search Count: 1,419
 |
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: J4U, PGT, PTY |
 |
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: J4U, PGT, PTY |
 |
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: PGT |
 |
Crystal Structure Of Human Programmed Cell Death 1 Ligand 1 (Pd-L1) Bound To Small Molecule Inhibitor Compound-10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: IMMUNE SYSTEM Ligands: A1L2P |
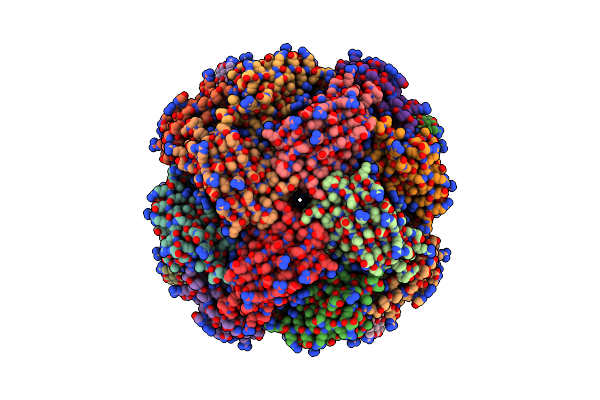 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
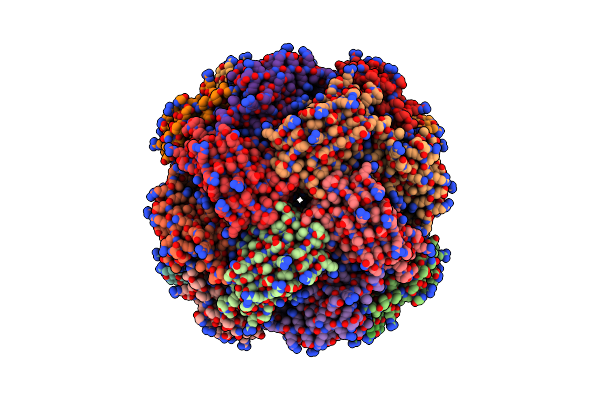 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, In Complex With Aminotriazole
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN, 3TR |
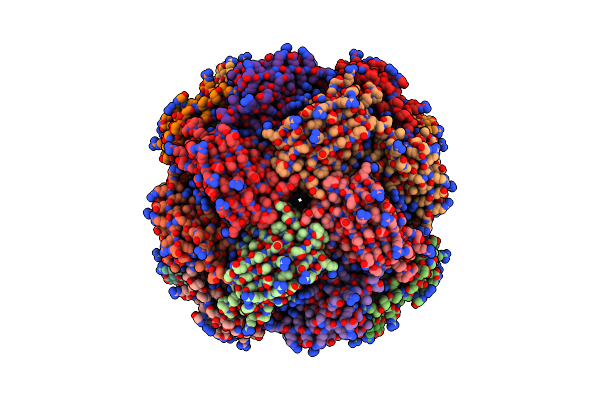 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
 |
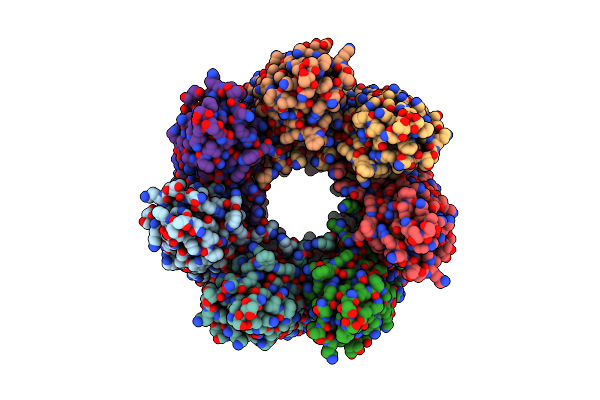
Alpha-Hemolysin Heptameric Late Pre-Pore State Derived From 10:0 Pc/Sphingomyelin Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN |
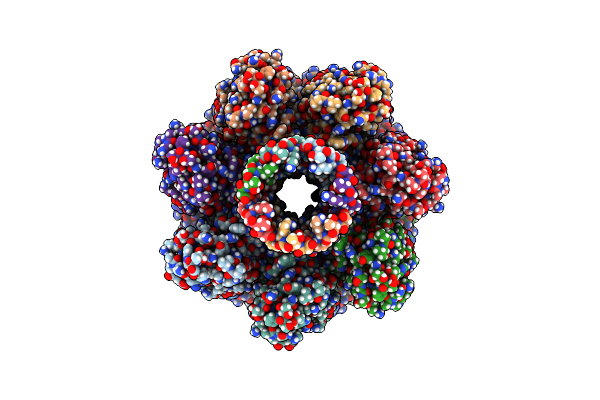 |
Alpha-Hemolysin Heptameric Pore State Derived From Egg Pc/Cholesterol (3:1 Molar Ratio) Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN |
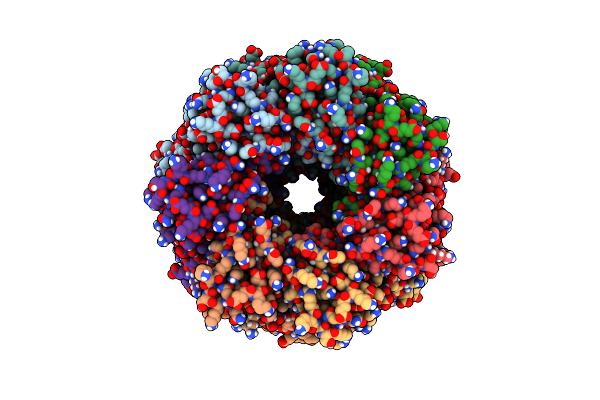 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN |
 |
Alpha-Hemolysin Heptameric Popc Bound Pore State Derived From Egg Pc/Cholesterol (3:1 Molar Ratio) Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: POV |
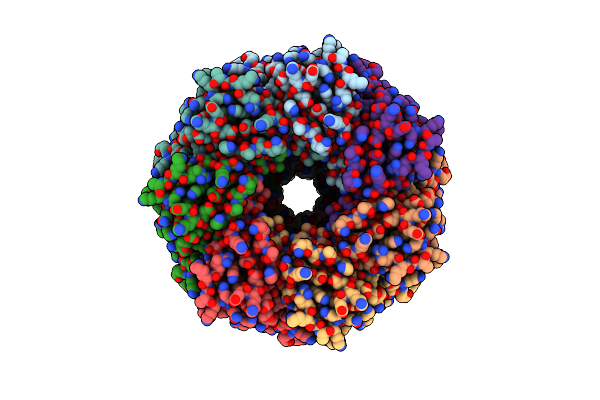 |
Alpha-Hemolysin Heptameric Pore State Bound To 10:0 Pc Lipid Chains Derived From 10:0 Pc Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: P1O |
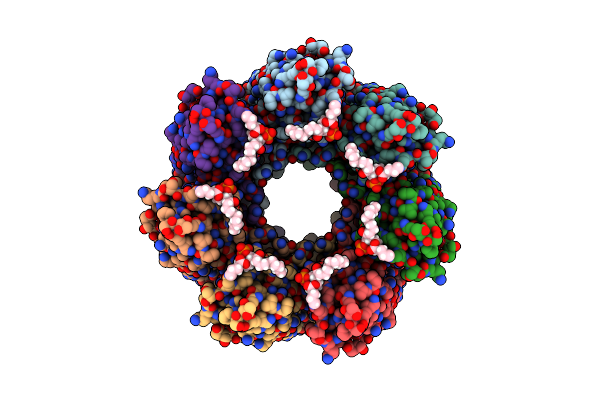 |
Alpha-Hemolysin Heptameric Pre-Pore State Bound To 10:Pc Lipid Chains Derived From 10:0 Pc Liposomes.
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: P1O |
 |
Alpha-Hemolysin Heptameric Late Pre-Pore State With Bound Lipids Derived From 10:0 Pc/Sphingomyelin Liposomes
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIPID BINDING PROTEIN Ligands: P1O |
 |
Organism: Xenopus laevis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-05-28 Classification: NUCLEAR PROTEIN Ligands: CL |
 |
Organism: Acinetobacter baumannii ab0057
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-03-19 Classification: LIGASE Ligands: PGE, GOL, MG, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: HYDROLASE Ligands: CL, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PROTEIN BINDING |

