IBIAP_1000000002

|
Indian major basmati paddy seed varieties images dataset |
The dataset contains images of 10 out of 32 notified Indian basmati seeds varieties (by the Government of India). Indian basmati paddy varieties included in the dataset are 1121, 1509, 1637, 1718, 1728, BAS-370, CSR 30, Type-3/Dehraduni Basmati, PB-1 and PB-6. Moreover, several images of other seeds and related entities available in the household have also been included in the dataset. Thus, the dataset contains 11 classes such that ten classes contain images from ten different basmati paddy varieties. In contrast, the 11th class- named “Unknown” contains images from a mixture of two morphologically similar paddy varieties (1121 and 1509), different pulses, other grains and related food entities. The Unknown class is useful in discriminating the paddy seeds from other types of seeds and related food entities. All the images were captured (in standard conditions) manually using an apparatus developed in-house and a tablet with a five-megapixel camera (5MP). The camera was used to capture 3210 RGB coloured images in JPG format. The data pre-processing was performed to generate the ready-to-use images for training and testing machine learning-based models. AI-based paddy seed variety classification models have been developed using the dataset. The dataset can be used to generate different types of AI-based models for adulteration detection, automated classification models (along with independent devices) at the time of rice threshing, and to increase the classification potential (Supplementing images representing additional basmati varieties).
|
PPS_1000000002
(Download Images)
(Print Records)
Views: 40
Images: 3210
Downloads: 85
Data size: 4.4GB
|
Indian major basmati paddy seed varieties images dataset |
Seeds from ten major Indian basmati paddy varieties were collected from the Indian Agricultural Research Institute (IARI), New Delhi, India. A total of 46 different types of pulses, grains and other food entities were also collected in-house to capture images other than paddy seeds. Moreover, a mixture of two morphologically similar paddy varieties (1121 and 1509) was also prepared in a separate vessel and its images constituted a new class (including pulses, grains and other food entities). Thus, the dataset (accessible on Mendeley) comprises 11 classes (10 basmati paddy seeds varieties and other grains and related food entities). An apparatus was designed to capture the images in standard conditions. A Micromax Canvas TAB P802 tablet, attached to the apparatus, was used to capture 3210 images.
|
https://www.sciencedirect.com/science/article/pii/S2352340920313421 |
International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi |
Open Access
|
Aug. 12, 2024 |
IBIAP_1000000003

|
An Opportunistic screening mammography dataset from a screening-naive population |
Mammographic images dataset from Indian population containing 1869 FFDM images and 1708 SM images, providing breast-level imaging data (BIRADS category and breast density) along with ground truth labels based on histopathology for cancers and follow-up scans for noncancers.
|
MAMOS_1000000004
(Download Images)
(Print Records)
Views: 83
Images: 3577
Downloads: 183
Data size: 63GB
|
An Opportunistic screening mammography dataset from a screening-naive population |
Mammographic dataset from Indian population containing 1869 FFDM images and 1708 SM images and provides breast-level imaging data (BIRADS category and breast density) along with ground truth labels based on histopathology for cancers and follow-up scans for noncancers.
|
N/A |
All India Institute of Medical Sciences (AIIMS), New Delhi |
Open Access
|
Sept. 10, 2024 |
IBIAP_1000000005

|
FruitNet: Indian fruits image dataset with quality for machine learning applications |
Fast and precise fruit classification or recognition as per quality parameter is the unmet need of agriculture business. This is an open research problem, which always attracts researchers. Machine learning and deep learning techniques have shown very promising results for the classification and object detection problems. Neat and clean dataset is the elementary requirement to build accurate and robust machine learning models for the real-time environment. With this objective we have created an image dataset of Indian fruits with quality parameter which are highly consumed or exported. Accordingly, we have considered six fruits namely apple, banana, guava, lime, orange, and pomegranate to create a dataset. The dataset is divided into three folders (1) Good quality fruits (2) Bad quality fruits, and (3) Mixed quality fruits each consists of six fruits subfolders. Total 19,500+ images in the processed format are available in the dataset. We strongly believe that the proposed dataset is very helpful for training, testing and validation of fruit classification or reorganization machine leaning model.
|
PPS_1000000009
(Download Images)
(Print Records)
Views: 16
Images: 19526
Downloads: 29
Data size: 3.1GB
|
FruitNet: Indian Fruits Dataset with quality (Good, Bad & Mixed quality) |
The profit percentage share of fruit market is substantial with respect to the total agriculture output. In the agro-industry fast and accurate fruit classification is the highest need. The fruits can be classified into different classes as per their external features like shape, size and color using some computer vision and deep learning techniques. High quality images of fruits are required to solve fruit classification and recognition problem. To build the machine learning models, neat and clean dataset is the elementary requirement. With this objective we have created the dataset of six popular Indian fruits named as “FruitNet”. This dataset consists of 19500+ high-quality images of 6 different classes of fruits in the processed format. The images are divided into 3 sub-folders 1) Good quality fruits 2) Bad quality fruits and 3) Mixed quality fruits. Each sub-folder contains the 6 fruits images i.e. apple, banana, guava, lime, orange, and pomegranate. Mobile phone with a high-end resolution camera was used to capture the images. The images were taken at the different backgrounds and in different lighting conditions. The proposed dataset can be used for training, testing and validation of fruit classification or reorganization model.
|
https://www.sciencedirect.com/science/article/pii/S2352340921009616 |
Vishwakarma University, Pune |
Open Access
|
Oct. 23, 2024 |
IBIAP_1000000007

|
Facilitating spice recognition and classification: An image dataset of Indian spices |
This data paper presents a comprehensive visual dataset of 19 distinct types of Indian spices, consisting of high-quality images meticulously curated to facilitate various research and educational applications. The dataset includes extensive imagery of the following spices: Asafoetida, Bay Leaf, Black Cardamom, Black Pepper, Caraway Seeds, Cinnamon Stick, Cloves, Coriander Seeds, Cubeb Pepper, Cumin Seeds, Dry Ginger, Dry Red Chilly, Fennel Seeds, Green Cardamom, Mace, Nutmeg, Poppy Seeds, Star Anise, and Stone Flowers. Each image in the dataset has been captured under controlled conditions to ensure consistency and clarity, making it an invaluable resource for studies in food science, agriculture, and culinary arts. The dataset can also support machine learning and computer vision applications, such as spice recognition and classification. By providing detailed visual documentation, this dataset aims to promote a deeper understanding and appreciation of the rich diversity of Indian spices.
|
PPS_1000000011
(Download Images)
(Print Records)
Views: 8
Images: 10991
Downloads: 32
Data size: 1.2GB
|
Facilitating spice recognition and classification: An image dataset of Indian spices |
This data paper presents a comprehensive visual dataset of 19 distinct types of Indian spices, consisting of high-quality images meticulously curated to facilitate various research and educational applications. The dataset includes extensive imagery of the following spices: Asafoetida, Bay Leaf, Black Cardamom, Black Pepper, Caraway Seeds, Cinnamon Stick, Cloves, Coriander Seeds, Cubeb Pepper, Cumin Seeds, Dry Ginger, Dry Red Chilly, Fennel Seeds, Green Cardamom, Mace, Nutmeg, Poppy Seeds, Star Anise, and Stone Flowers. Each image in the dataset has been captured under controlled conditions to ensure consistency and clarity, making it an invaluable resource for studies in food science, agriculture, and culinary arts. The dataset can also support machine learning and computer vision applications, such as spice recognition and classification. By providing detailed visual documentation, this dataset aims to promote a deeper understanding and appreciation of the rich diversity of Indian spices.
|
https://doi.org/10.1016/j.dib.2024.110936 |
Vishwakarma University, Pune |
Open Access
|
Dec. 12, 2024 |
IBIAP_1000000008

|
PSFD-Musa: A dataset of banana plant, stem, fruit, leaf, and disease |
Varieties of banana plants can be found worldwide. It grows in Tropical regions and requires a hot and humid climate to develop itself. It is seen that each part of a banana plant can get infected with different types of bacterial, fungal, and viral diseases. Out of which many of them are dangerous diseases that affect it and its production. Deficiency diseases too can incur a heavy loss over the banana plantations. To get familiarized with different varieties of banana plants and to know some of the common diseases that affect the plants, we have created a PSFD-Musa DATASET, for the banana plants that are indigenously found in different parts of Assam. The dataset is divided into 3 subfolders. The first folder comprises the images of different varieties of banana plants which further consists of 7 classes namely Malbhog fruit (Musa assamica), Malbhog leaf (Musa assamica), Jahaji fruit (Musa chinensis), Jahaji stem (Musa chinensis), Jahaji leaf (Musa chinensis), Kachkol fruit (Musa paradisiaca L.), Bhimkol leaf (M. Balbisiana Colla). The second folder comprises different diseases that affect the banana plants which again comprises 7 classes namely: Bacterial Soft Rot, Banana Fruit Scarring Beetle, Black Sigatoka, Yellow Sigatoka, Panama disease, Banana Aphids, and PseudoStem Weevil. And the last folder is of the deficiencies that hamper the plants which are of 1 class namely: Potassium deficiency. The images provided here are raw as well as processed data and are in the format of .jpg.
|
PPS_1000000012
(Download Images)
(Print Records)
Views: 9
Images: 9463
Downloads: 32
Data size: 109MB
|
PSFD-Musa: A dataset of banana plant, stem, fruit, leaf, and disease |
In recent times, the classification and identification of different fruits and food crops have become a necessity in the field of agricultural science; for sustainable growth. Probable processes have been developed worldwide to improve the production of food crops. Problem-specific, clean and crisp datasets are also lagging in the sector. This article introduces an image dataset of varieties of banana plants and the diseases related to them. The varieties of Banana plants that we have considered in the dataset are the Malbhog (Musa assamica), Jahaji (Musa chinensis), Kachkol (Musa paradisiaca L.), Bhimkol (M. Balbisiana Colla). And the diseases and pathogens that we have considered here are the Bacterial Soft Rot, Banana Fruit Scarring Beetle, Black Sigatoka, Yellow Sigatoka, Panama disease, Banana Aphids, and Pseudo-Stem Weevil. A dataset of Potassium deficiency has been also considered in this article. A total of 8000+ processed images are present in the dataset. The purpose of this article is to provide the Researchers and Students in getting access to our dataset that would help them in their research and in developing some machine learning models.
|
https://www.sciencedirect.com/science/article/pii/S2352340922006242 |
Gauhati University, Assam |
Open Access
|
Dec. 19, 2024 |
IBIAP_1000000009

|
High-resolution AI image dataset for diagnosing oral submucous fibrosis and squamous cell carcinoma |
Oral cancer is a global health challenge with a difficult histopathological diagnosis. The accurate histopathological interpretation of oral cancer tissue samples remains difficult. However, early diagnosis is very challenging due to a lack of experienced pathologists and inter- observer variability in diagnosis. The application of artificial intelligence (deep learning algorithms) for oral cancer histology images is very promising for rapid diagnosis. However, it requires a quality annotated dataset to build AI models. We present ORCHID (ORal Cancer Histology Image Database), a specialized database generated to advance research in AI-based histology image analytics of oral cancer and precancer. The ORCHID database is an extensive multicenter collection of high-resolution images captured at 1000X effective magnification (100X objective lens), encapsulating various oral cancer and precancer categories, such as oral submucous fibrosis (OSMF) and oral squamous cell carcinoma (OSCC). Additionally, it also contains grade-level sub-classifications for OSCC, such as well- differentiated (WD), moderately-differentiated (MD), and poorly-differentiated (PD). The database seeks to aid in developing innovative artificial intelligence-based rapid diagnostics for OSMF and OSCC, along with subtypes.
|
HISTOS_1000000013
(Download Images)
(Print Records)
Views: 26
Images: 14705
Downloads: 75
Data size: 54GB
|
High-resolution AI image dataset for diagnosing oral submucous fibrosis and squamous cell carcinoma |
The number of images available in each of the five classes(folders), which are as follows, Normal, OSMF, WDOSCC, MDOSCC, and PDOSCC. Each class folder consists of subfolders representing different tissue slides collected from different patients. We have made an initial attempt to provide a comprehensive image database for two of the most prominent oral conditions, OSCC and OSMF. We believe that more such databases will be made publicly available in the near future. These comprehensive image databases will facilitate the development of accurate AI-based diagnostic tools for oral diseases, ultimately improving patient care and outcomes in the field of oral healthcare. In future, integration of databases comprising molecular markers, transcriptome, metabolome, and other biomarkers, combined with oral histological image through advanced AI-driven imaging techniques, holds great promise in improving diagnostic accuracy and precision. This potential has already been observed in the diagnosis of lung and breast cancers. This expansion will aid in developing a more comprehensive AI-driven diagnostic tool.
|
https://doi.org/10.1038/s41597-024-03836-6 |
Jamia Millia Islamia, Delhi |
Open Access
|
Jan. 13, 2025 |
IBIAP_1000000010

|
Histopathological images of thyroid lesions |
The dataset comprises 154,498 images derived from 134 slides, representing 125 thyroid nodules from 118 patients. These images encompass six types of thyroid lesions: NIFTP, HTN, FA, IEFVPTC, IFSPTC, and CPTC.
|
HISTOS_1000000014
(Download Images)
(Print Records)
Views: 30
Images: 154498
Downloads: 36
Data size: 198GB
|
Utility of Artificial Intelligence in differentiating Non- invasive Follicular Thyroid Neoplasm with Papillary like Nuclear Features from other follicular-patterned thyroid benign and malignant lesions |
The introduction of the term non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) in 2016 marked a pivotal shift in the classification of encapsulated follicular variants of papillary thyroid carcinoma (eFVPTC) lacking invasive features. While this reclassification significantly reduced overtreatment, the histopathological diagnosis of NIFTP remains challenging due to overlapping features with other thyroid lesions and inter-observer variability. This study presents a novel deep learning (DL)-based, three-stage diagnostic pipeline for distinguishing NIFTP from a wide spectrum of thyroid lesions, including benign and malignant mimics. By replicating the diagnostic strategy of histopathologists, the algorithm evaluates architectural patterns and nuclear features with high precision. Our approach has a potential to enhance diagnostic accuracy in a cost-effective and scalable manner, complementing existing diagnostic methods and thus optimizing clinical decision-making and improving the management of patients with thyroid neoplasms.
|
https://link.springer.com/article/10.1007/s12022-025-09865-0 |
- All India Institute of Medical Sciences (AIIMS), New Delhi
- International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi
|
Open Access
|
June 2, 2025 |
IBIAP_1000000011

|
Chákṣu: A glaucoma specific fundus image database |
We introduce Chákṣu–a retinal fundus image database for the evaluation of computer-assisted glaucoma prescreening techniques. The database contains 1345 color fundus images acquired using three brands of commercially available fundus cameras. Each image is provided with the outlines for the optic disc (OD) and optic cup (OC) using smooth closed contours and a decision of normal versus glaucomatous by five expert ophthalmologists. In addition, segmentation ground-truths of the OD and OC are provided by fusing the expert annotations using the mean, median, majority, and Simultaneous Truth and Performance Level Estimation (STAPLE) algorithm. The performance indices show that the ground-truth agreement with the experts is the best with STAPLE algorithm, followed by majority, median, and mean. The vertical, horizontal, and area cup-to-disc ratios are provided based on the expert annotations. Image-wise glaucoma decisions are also provided based on majority voting among the experts. Chákṣu is the largest Indian-ethnicity-specific fundus image database with expert annotations and would aid in the development of artificial intelligence based glaucoma diagnostics.
|
OPTHS_1000000015
(Download Images)
(Print Records)
Views: 13
Images: 1345
Downloads: 18
Data size: 11GB
|
Chákṣu: A glaucoma specific fundus image database |
Glaucoma is a chronic, irreversible, and slowly progressing optical neuropathy that damages the optic nerve. Depending on the extent of damage to the optic nerve, glaucoma can cause moderate to severe vision loss. Glaucoma is asymptomatic in the early stages. It is not curable, and the lost vision cannot be restored. However, by early screening and detection, the progression of the disease could be slowed down. Color fundus imaging is the most viable non-invasive means of examining the retina for glaucoma. The widest application of fundus imaging is in optic nerve head or optic disc examination for glaucoma management. Fundus imaging is widely used due to the relative ease of establishing a digital baseline for assessing the progression of the disease and the effectiveness of the treatment. Fundus imaging technology is developing rapidly and several exciting products with fully automated software applications for retinal disease diagnosis are on the horizon. State-of-the-art tools based on image processing and deep learning algorithms are becoming increasingly useful and relevant. However, before deploying them in a clinical setting, a thorough validation over benchmark datasets is essential. The development of a large database with multiple expert annotations is a laborious and tedious task. A large annotated glaucoma-specific fundus image database is lacking, which is a gap that the Chákṣu database reported in this paper attempts to fill. Several retinal fundus image databases are publicly available to facilitate research and performance comparison of segmentation and classification algorithms.
|
https://doi: 10.1038/s41597-023-01943-4 |
Indian Institute of Science, Bangalore |
Open Access
|
March 16, 2025 |
IBIAP_1000000012

|
Histopathological imaging database for oral cancer analysis |
The repository is composed of 1224 images divided into two sets of images with two different resolutions. First set consists of 89 histopathological images with the normal epithelium of the oral cavity and 439 images of Oral Squamous Cell Carcinoma (OSCC) in 100x magnification. The second set consists of 201 images with the normal epithelium of the oral cavity and 495 histopathological images of OSCC in 400x magnification. The images were captured using a Leica ICC50 HD microscope from Hematoxyline and Eosin (H&E) stained tissue slides collected, prepared and catalogued by medical experts from 230 patients. A subset of 269 images from the second data set was used to detect OSCC based on textural features. Histopathology plays a very important role in diagnosing a disease. It is the investigation of biological tissues to detect the presence of diseased cells in microscopic detail. It usually involves a biopsy. Till date biopsy is the gold-standard test to diagnose cancer. The biopsy slides are examined based on various cytological criteria under a microscope. Therefore, there is a high possibility of not retaining uniformity and ensuring reproducibility in outcomes. Computational diagnostic tools, on the other hand, facilitate objective judgments by making the use of the quantitative measure. This dataset can be utilized in establishing automated diagnostic tool using Artificial Intelligence approaches.
|
HISTOS_1000000016
(Download Images)
(Print Records)
Views: 23
Images: 1224
Downloads: 29
Data size: 2.9GB
|
Histopathological imaging database for oral cancer analysis |
This is the first dataset containing histopathological images of the normal epithelium of the oral cavity and OSCC. The images were captured using a Leica ICC50 HD microscope from Hematoxyline and Eosin (H&E) stained tissue slides collected, prepared and catalogued by medical experts from 230 patients. Invasion of the tumour into the basement membrane is a very important architectural feature for diagnosing OSCC. Researchers can use 100x magnified images for architectural or tissue level analysis. These can also be used in feature extraction like shape, texture or colour feature extraction, segmentation of the epithelial layer, invasion of tumour into the basement membrane, or in categorizing images in normal and malignant category considering the whole architecture of the images. 400x magnified images can be used for tissue level analysis, such as in the automated diagnosis of the disease based on the textural feature. This dataset can be utilized in establishing automated diagnostic tool using Artificial Intelligence approaches. These data can be used as a gold standard for histopathological analysis of OSCC. This dataset can be used for a comparative evaluation of one's experimental findings in future when more dataset of such kind is available.
|
https://doi.org/10.1016/j.dib.2020.105114 |
Institute of Advanced Study in Science and Technology, Guwahati, Assam |
Open Access
|
March 24, 2025 |
IBIAP_1000000016

|
Dry fruit image dataset for machine learning applications |
The "Dry Fruit Image Dataset" is a collection of 11500+ processed high-quality images representing 12 distinct classes of dry fruits. The 4 dry fruits—Almonds, Cashew Nuts, Raisins, and Dried Figs (Anjeer)—along with 3 sub-types of each are contained in the sub-folders, making a total of 12 distinct classes. These pictures were taken with a high-definition camera on cell phones. The dataset contains images in different lighting conditions as well as with different backgrounds. This dataset can be used for building machine learning models for the classification and recognition of Dry Fruits, requiring neat, appropriately tagged, and high-quality images. The dry fruit classification algorithm can be trained, tested, and validated using this dataset. Furthermore, it is beneficial for dry fruit research, education, and medicinal purposes.
|
PPS_1000000020
(Download Images)
(Print Records)
Views: 7
Images: 11520
Downloads: 10
Data size: 323MB
|
Dry fruit image dataset for machine learning applications |
Dry fruits are convenient and nutritious snacks that can provide numerous health benefits. They are packed with vitamins, minerals, and fibres, which can help improve overall health, lower cholesterol levels, and reduce the risk of heart disease. Due to their health benefits, dry fruits are an essential part of a healthy diet. In addition to health advantage, dry fruits have high commercial worth. The value of the global dry fruit market is estimated to be USD 6.2 billion in 2021 and USD 7.7 billion by 2028. The appearance of dry fruits is utilized for assessing their quality to a great extent, requiring neat, appropriately tagged, and high-quality images. Hence, this dataset is a valuable resource for the classification and recognition of dry fruits. With over 11500+ high-quality processed images representing 12 distinct classes, this dataset is a comprehensive collection of different varieties of dry fruits. The four dry fruits included in this dataset are Almonds, Cashew Nuts, Raisins, and Dried Figs (Anjeer), along with three subtypes of each. This makes it a total of 12 distinct classes of dry fruits, each with its unique features, shape, and size. The dataset will be useful for building machine learning models that can classify and recognize different types of dry fruits under different conditions, and can also be beneficial for dry fruit research, education, and medicinal purposes.
Due to their nutritional value and health advantages, dry fruits have been consumed for a very long time. One of the best strategies to improve general health is to include dry fruits in the diet.
|
https://doi.org/10.1016/j.dib.2023.109325 |
Vishwakarma Institute of Information Technology, Pune |
Open Access
|
April 24, 2025 |
IBIAP_1000000015
|
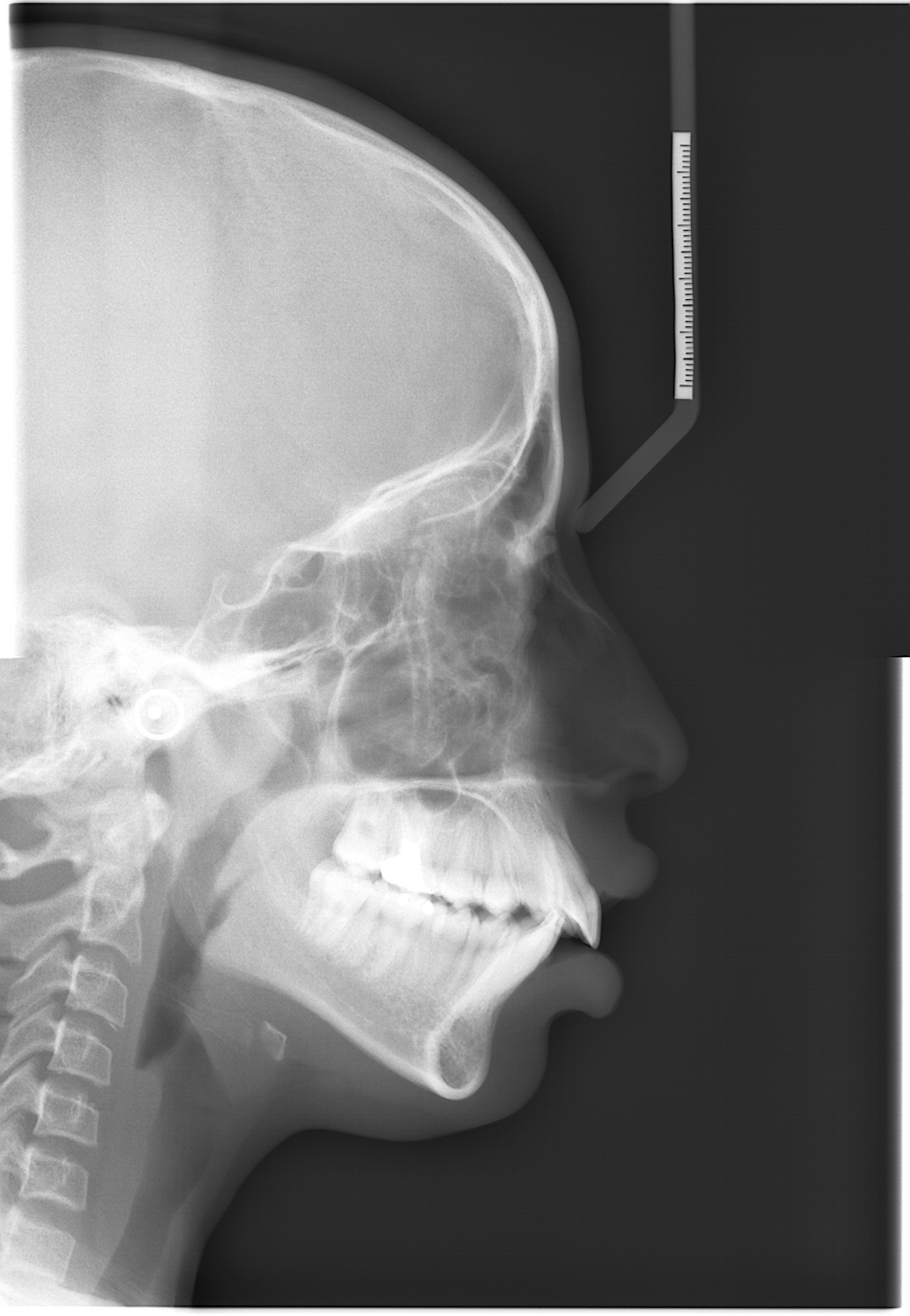
DiverseCeph19: A Cephalometric Landmarks Annotation Dataset |
Automation of cephalometric landmark annotation holds significant importance within the domain as a research area. This article presents a dataset consisting of 1692 cephalometric radiograph images, along with manually annotated data on 19 anatomical cephalometric landmarks. The data is available in individual text files corresponding to individual radiographic image files. Along with this, the latest annotation formats like COCO JSON and YOLO file formats for artificial intelligence (AI) model training are also available. Annotations are performed by experienced medical professionals with more than twenty years of experience and technical professionals with 6 years of research experience in the domain. The images are preprocessed and segregated based on resolution, image quality, dental structure, and other artifacts. The final image data is saved in BMP format with 0.127 mm/pixel resolution. The images refer to cephalometric analysis performed between 2011 and 2022 in patients treated at JSS Dental College and Hospital (JSS DCH), with patients age between 5 to 60 years. The demographic information of patients is not available with individual image data. Ethical clearance is obtained from the Institutional Ethical Committee of JSS DCH. Various levels of experimentation are carried out using the dataset, and the results demonstrate robust performance. Landmark annotation based on segregation type is one of the first types in this area of cephalometric landmark annotation. The availability of this dataset offers researchers a robust platform for investigating and conducting experiments using machine learning and deep learning techniques.
|
XRS_1000000021
(Request Access)
Views: 0
Images: 1692
Downloads: 0
Data size: N/A
|
Automation of Cephalometric Landmarks Annotation |
Each image collected from the hospital is first cropped to eliminate the patient-related information like name, sex, age, and date of image capture. Images are cropped to 2 distinct resolutions: 1341x1938 and 1257x1672 based on the lateral cephalometric region covered in the raw images and are saved in BMP format. Further, within individual resolution, images are segregated based on various factors, like 1. Teeth structure (supernumerary teeth, unerupted teeth) 2. Visibility of soft tissue profile. 3. Presence of orthodontic braces 4. Presence of earrings or a nose pin. Segregated image data is annotated by technical and medical professionals using ImageJ software (v1.53q). Identified landmarks are reviewed by experienced medical professionals, and finally, the landmark pixel locations are extracted into a text file.
|
https://doi.org/10.1016/j.compbiomed.2024.109318 |
JSS Science and Technology University, Mysuru |
Managed Access
|
May 6, 2026 |
IBIAP_1000000017

|
Enhanced deep learning technique for sugarcane leaf disease classification and mobile application integration |
With an emphasis on classifying diseases of sugarcane leaves, this research suggests an attention-based multilevel deep learning architecture for reliably classifying plant diseases. The suggested architecture comprises spatial and channel attention for saliency detection and blends features from lower to higher levels. On a self-created database, the model outperformed cutting-edge models like VGG19, ResNet50, XceptionNet, and EfficientNet_B7 with an accuracy of 86.53%. The findings show how essential all-level characteristics are for categorizing images and how they can improve efficiency even with tiny databases. The suggested architecture has the potential to support the early detection and diagnosis of plant diseases, enabling fast crop damage mitigation. Additionally, the implementation of the proposed AMRCNN model in the Android phone-based application gives an opportunity for the widespread use of mobile phones in the classification of sugarcane diseases.
|
PPS_1000000022
(Download Images)
(Print Records)
Views: 6
Images: 2521
Downloads: 12
Data size: 160MB
|
Sugarcane leaf disease dataset |
Manually collected image dataset of sugarcane leaf disease. It has mainly five categories in it. Healthy, Mosaic, Redrot, Rust and Yellow disease. The dataset has been captured with smart phones of various configuration to maintain the diversity. It contains total 2569 images including all categories. This database has been collected in Maharashtra, India. The database is balanced and contains good variety. The image sizes are not constant as it originates form various capturing devices. All images are in RGB format.
|
https://doi.org/10.1016/j.heliyon.2024.e29438 |
G. H. Raisoni College of Engineering & Management, Wagholi, Pune, Maharashtra |
Open Access
|
June 24, 2025 |
IBIAP_1000000018

|
Sugarcane leaf dataset: A dataset for disease detection and classification for machine learning applications |
Sugarcane, a vital crop for the global sugar industry, is susceptible to various diseases that significantly impact its yield and quality. Accurate and timely disease detection is crucial for effective management and prevention strategies. We persent the “Sugarcane Leaf Dataset" consisting of 6748 high-resolution leaf images classified into nine disease categories, a healthy leaves category, and a dried leaves category. The dataset covers diseases such as smut, yellow leaf disease, pokkah boeng, mosale, grassy shoot, brown spot, brown rust, banded cholorsis, and sett rot. The dataset's potential for reuse is significant. The provided dataset serves as a valuable resource for researchers and practitioners interested in developing machine learning algorithms for disease detection and classification in sugarcane leaves. By leveraging this dataset, various machine learning techniques can be applied, including deep learning, feature extraction, and pattern recognition, to enhance the accuracy and efficiency of automated sugarcane disease identification systems. The open availability of this dataset encourages collaboration within the scientific community, expediting research on disease control strategies and improving sugarcane production. By leveraging the “Sugarcane Leaf Dataset,” we can advance disease detection, monitoring, and management in sugarcane cultivation, leading to enhanced agricultural practices and higher crop yields.
|
PPS_1000000023
(Download Images)
(Print Records)
Views: 5
Images: 6748
Downloads: 9
Data size: 746MB
|
Sugarcane leaf dataset |
This Sugarcane Leaf Dataset contains a diverse collection of 6748 high-resolution images of sugarcane leaves. The images are stored in JPEG format and have dimensions of 768 × 1024 pixels. The dataset is categorized into 11 distinct classes, including nine disease categories, a healthy leaves category, and a dried leaves category. The disease categories cover a range of common sugarcane leaf diseases, such as smut, yellow leaf disease, pokkah boeng, mosale, grassy shoot, brown spot, brown rust, banded cholorsis, and sett rot. Each category is labelled and organized in separate folders, ensuring easy access and identification of specific disease samples. The images were collected through extensive field surveys conducted in sugarcane-growing regions. The data collection process involved using quality cameras to capture images from various angles, including both sides of the leaves. Images were taken in the field and by cutting/separating individual leaves, capturing different stages and manifestations of the diseases. This approach ensures a comprehensive representation of the visual characteristics of sugarcane leaf diseases within the dataset. The dataset's images are of high quality, with a resolution set at 72 dots per inch (dpi), ensuring clear and detailed visual representation of the sugarcane leaf samples.
|
https://doi.org/10.1016/j.dib.2024.110268 |
Vishwakarma University, Pune |
Open Access
|
July 16, 2025 |
IBIAP_1000000021
|
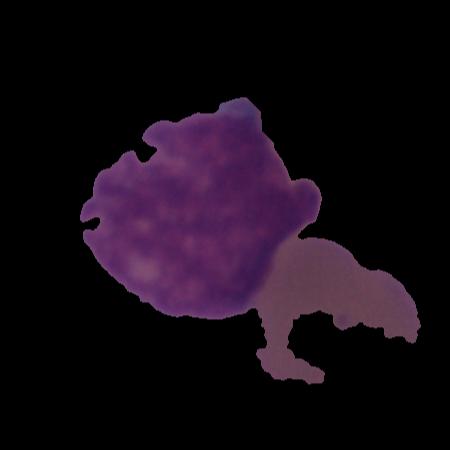
C-NMC: B-lineage acute lymphoblastic leukaemia (B-ALL): A blood cancer dataset |
Development of computer-aided cancer diagnostic tools is an active research area owing to the advancements in deep-learning domain. Such technological solutions provide affordable and easily deployable diagnostic tools. Leukaemia, or blood cancer, is one of the leading cancers causing more than 0.3 million deaths every year. In order to aid the development of such an AI-enabled tool, we collected and curated a microscopic image dataset, namely C-NMC, of more than 15000 cancer cell images at a very high resolution of B-Lineage Acute Lymphoblastic Leukaemia (B-ALL). The dataset is prepared at the subject-level and contains images of both healthy and cancer patients. So far, this is the largest (as well as curated) dataset on B-ALL cancer in the public domain. C-NMC is also available at The Cancer Imaging Archive (TCIA), USA and can be helpful for the research community worldwide for the development of B-ALL cancer diagnostic tools. This dataset was utilized in an international medical imaging challenge held at ISBI 2019 conference in Venice, Italy. In the published article, we have presented a detailed description and challenges of this dataset. We have also presented benchmarking results of all the methods applied so far on this dataset.
|
HISTOS_1000000025
(Download Images)
(Print Records)
Views: 7
Images: 15114
Downloads: 6
Data size: 859MB
|
An image dataset of B-lineage acute lymphoblastic leukaemia (B-ALL) and healthy hematogones |
This study provides a dataset of white blood cancer, namely, B-Lineage Acute Lymphoblastic Leukaemia (B-ALL) along with the healthy hematogones. The dataset has been split at the subject-level into the training and the test sets. Specifically, the training set contains 12528 cell images of 8491 cancer lymphoblasts and 4037 healthy blasts (also called as hematogones). Cancer cells belong to 60 cancer patients, while normal cells belong to 41 subjects. The test set contains 2586 cell images belonging to 8 healthy (or control) subjects and 9 cancer patients. The training and test set are distributed such that there are no common subjects between the two sets. This dataset was released during the IEEE ISBI 2019 medical imaging challenge in three phases: 1) initial train phase, 2) preliminary test phase, and 3) final test phase. In the initial train phase, the dataset was released for all the registered participants for training their classification networks. In the preliminary test phase, a preliminary test set was released to allow the testing of the performance of the participants’ models. The top participants in this phase were shortlisted to move to the next phase of the challenge and were also provided the ground truth (GT) of the preliminary test set for improving the performance in the next round. Hence, the participants had the GT of the initial training data and the preliminary stage’s test data. Together, this data was used by the participants for the training of their models and tested on the final test data released in the final test phase to decide the ranking on the leaderboard. The dataset arranged in these three phases was accordingly released publicly for use by the future researchers. The GT of training and preliminary test data is released, while those of test data have not been released.
|
https://doi.org/10.1016/j.medengphy.2022.103793 |
- All India Institute of Medical Sciences (AIIMS), New Delhi
- Indraprastha Institute of Information Technology, Delhi
|
Open Access
|
Aug. 25, 2025 |
IBIAP_1000000023
|
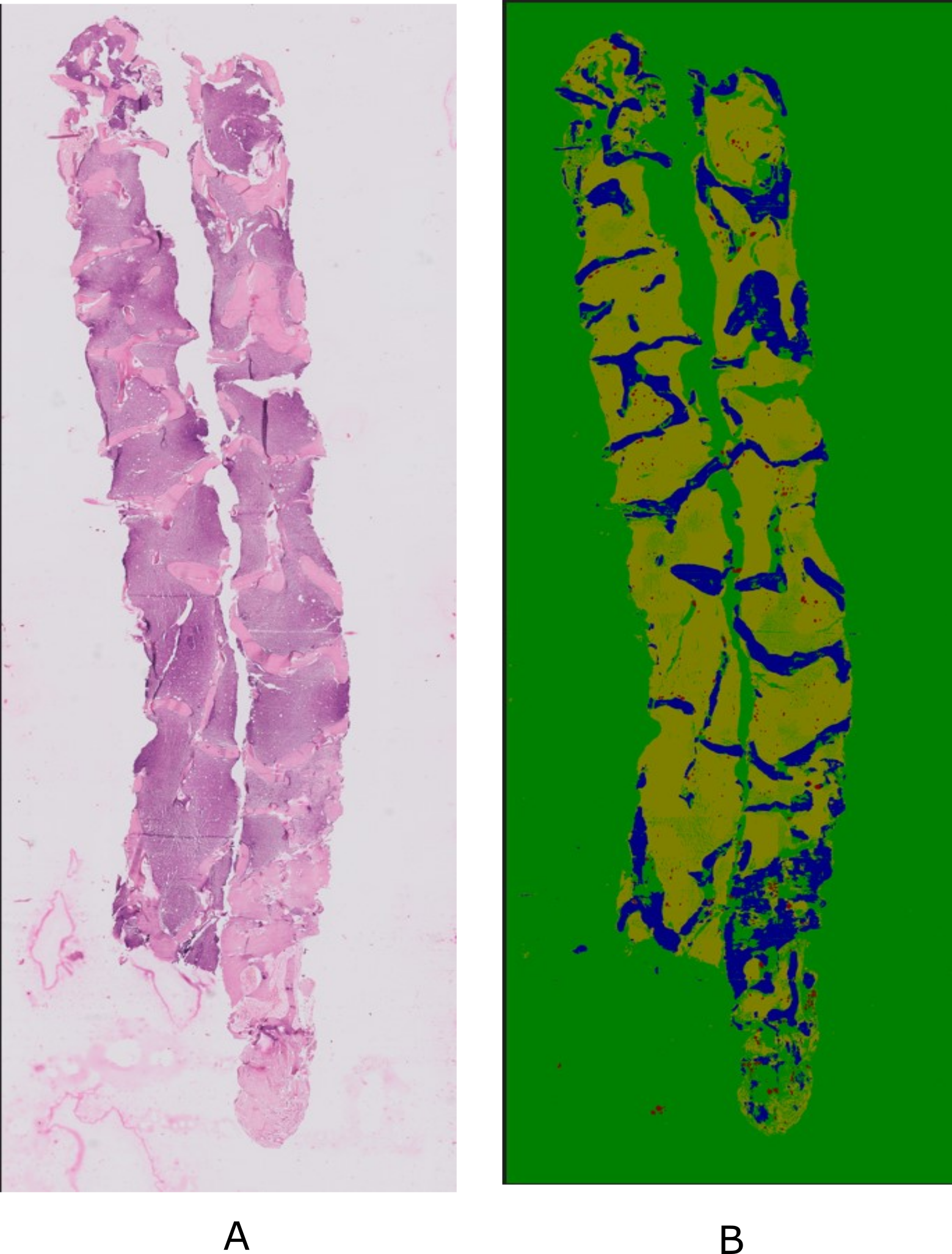
Developing a Deep Learning Framework for Clinical Use in Estimating Cellularity in Bone Marrow Biopsy Specimens |
Bone marrow examination has become increasingly important for the diagnosis and treatment of hematologic and other illnesses. The present methods for analyzing bone marrow biopsy samples involve subjective and inaccurate assessments by visual estimation by pathologists. Thus, there is a need to develop automated tools to assist in the analysis of bone marrow samples. However, there is a lack of publicly available standardized and high-quality datasets that can aid in the research and development of automated tools that can provide consistent and objective measurements. In this paper, we present a comprehensive Bone Marrow Biopsy (BaMBo) dataset consisting 229 semantic-segmented bone marrow biopsy images, specifically designed for the automated calculation of bone marrow cellularity. Our dataset comprises high-resolution, generalized images of bone marrow biopsies, each annotated with precise semantic segmentation of different haematological components. These components are divided into 4 classes: Bony trabeculae, adipocytes, cellular region and Background. The annotations were performed with the help of two experienced hematopathologists that were supported by state-of-the-art DL models and image processing techniques.
|
HISTOS_1000000027
(Download Images)
(Print Records)
Views: 3
Images: 229
Downloads: 6
Data size: 545MB
|
BaMBo: An Annotated Bone Marrow Biopsy Dataset for Segmentation Task |
Data collection involved the acquisition of bone marrow trephine (BMT) biopsy specimens using standardized procedures at Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh. These procedures, as described later, are medical standards. The dataset comprises 229 biopsy images from a total of 38 patients, diagnosed with varied hematological states covering myeloproliferative neoplasms, leukemia, aplastic anemia, uninvolved staging marrows among others. Subjects were chosen at random from both the genders, contains a large age gap and varied medical history. All the patient related identifiers were removed from the dataset and the dataset was completely anonymized for patient confidentiality.
|
Anilpreet Singh, Satyender Dharamdasani, Praveen Sharma, Sukrit Gupta. BaMBo: An Annotated Bone Marrow Biopsy Dataset for Segmentation Task, Open Data Workshop, MICCAI 2024 |
- Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh
- Indian Institute of Technology Ropar, Rupnagar
|
Open Access
|
Oct. 16, 2025 |
IBIAP_1000000024
|
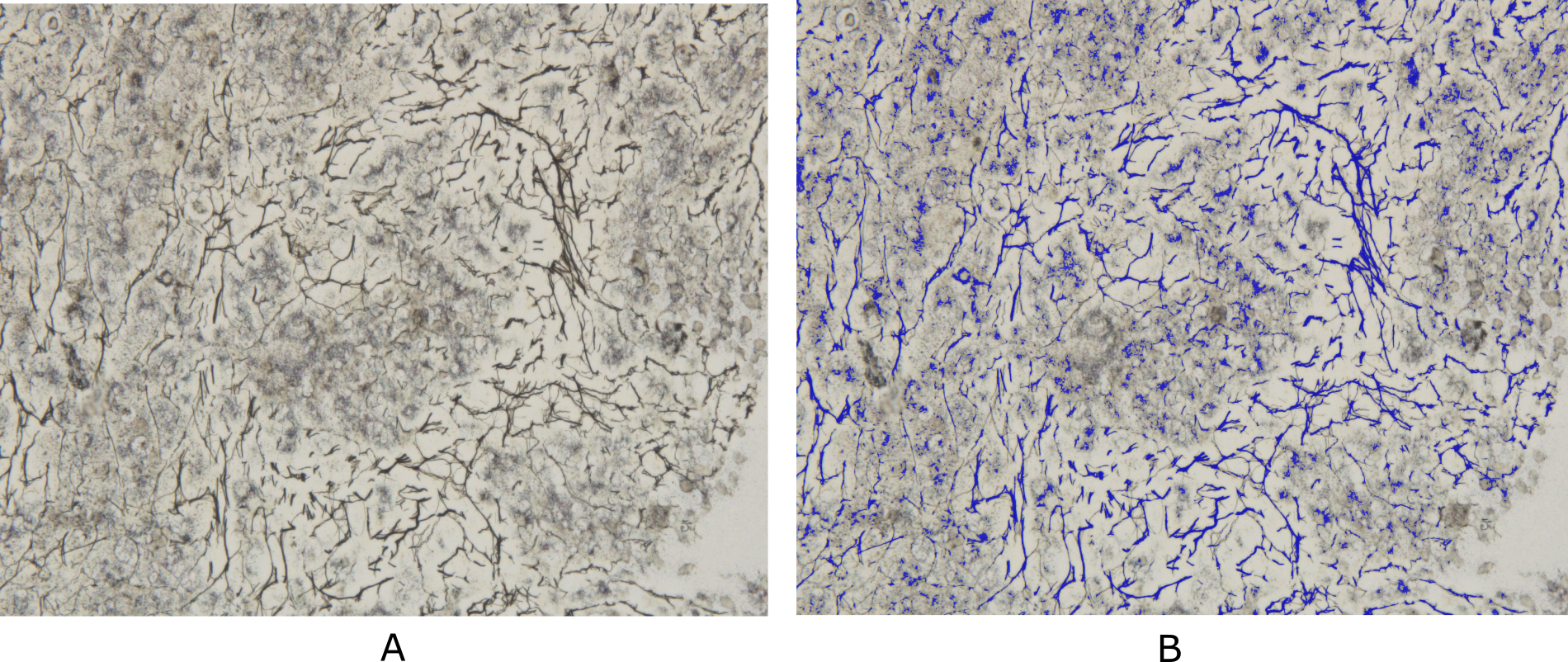
Developing Deep Learning Models for Quantitative Estimation of Reticulin Fibers in Bone Marrow Trephine Biopsy Specimens |
Bone marrow reticulin fibrosis is associated with varied benign as well as malignant hematological conditions. The assessment of reticulin fibrosis is important in the diagnosis, prognostication and management of such disorders. The current methods for quantification of reticulin fibrosis are inefficient and prone to errors. Therefore, there is a need for automated tools for accurate and consistent quantification of reticulin. However, the lack of standardized datasets has hindered the development of such tools. In this study, we present a comprehensive dataset that comprises of 250 Bone Marrow Biopsy images for Reticulin (BoMBR) quantification. These images were meticulously annotated for semantic segmentation, with the focus on performing reticulin fiber quantification. This annotation was done by two trained hematopathologists who were aided by Deep Learning (DL) models and image processing techniques that generated a rough automated annotation for them to start with. This ensured precise delineation of the reticulin fibers alongside other cellular components such as bony trabeculae, fat, and cells. This is the first publicly available dataset in this domain with the aim to catalyze advancements the development of computational models for improved reticulin quantification.
|
HISTOS_1000000028
(Download Images)
(Print Records)
Views: 8
Images: 250
Downloads: 4
Data size: 996MB
|
BoMBR: Bone Marrow Biopsy Dataset for Segmentation of Reticulin Fibers |
We propose the BoMBR dataset containing 250 Bone Marrow Trephine (BMT) pixel-wise annotated images. Besides the pixel-wise annotation masks, we give information regarding the grade of Marrow Fibrosis, percentage cellular area covered by reticulin, and the average value of the elongation factor of the reticulin fibers in the image. The percentage of cellular area covered by reticulin indicates differences in the amount of area covered by reticulin fibers across different grades, while the elongation factor helps understand changes in the shape of reticulin fibers as the grade increases. This is by far the first such publicly available resource for annotated BMT images that focuses on reticulin fibrosis both globally and in the context of the Indian subcontinent. Our dataset aims to facilitate the effective and objective quantitative measurement of reticulin fibers, moving beyond the current qualitative, observer-dependent scoring systems. Beyond its primary application in the grading of myelofibrosis, the dataset may also serve as a benchmark for further studies. This includes its potential to assist hematopathologists in the classification of Myeloproliferative Neoplasms, assessment of disease progression, and quantitative monitoring of the effects of myelofibrosis reversal in patients, particularly in the context of clinical trials and novel targeted therapies. The annotated dataset of BMT biopsy images presented in this study provides a valuable resource for quantitative assessment of fibrosis severity in hematological disorders. By leveraging automated segmentation followed by expert annotation, we have created a comprehensive dataset covering a range of Marrow Fibrosis grades (MF-0 to MF-3). This dataset includes detailed annotations for reticulin fibers, bony trabeculae, fat regions, and cellular region, facilitating in-depth analysis of bone marrow pathology.
|
Panav Raina, Satyender Dharamdasani, Dheeraj Chinnam, Praveen Sharma, Sukrit Gupta BoMBR: An Annotated Bone Marrow Biopsy Dataset for Segmentation of Reticulin Fibers Open Data @ MICCAI 2024 |
- Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh
- Indian Institute of Technology Ropar, Rupnagar
|
Open Access
|
Sept. 23, 2025 |
IBIAP_1000000020
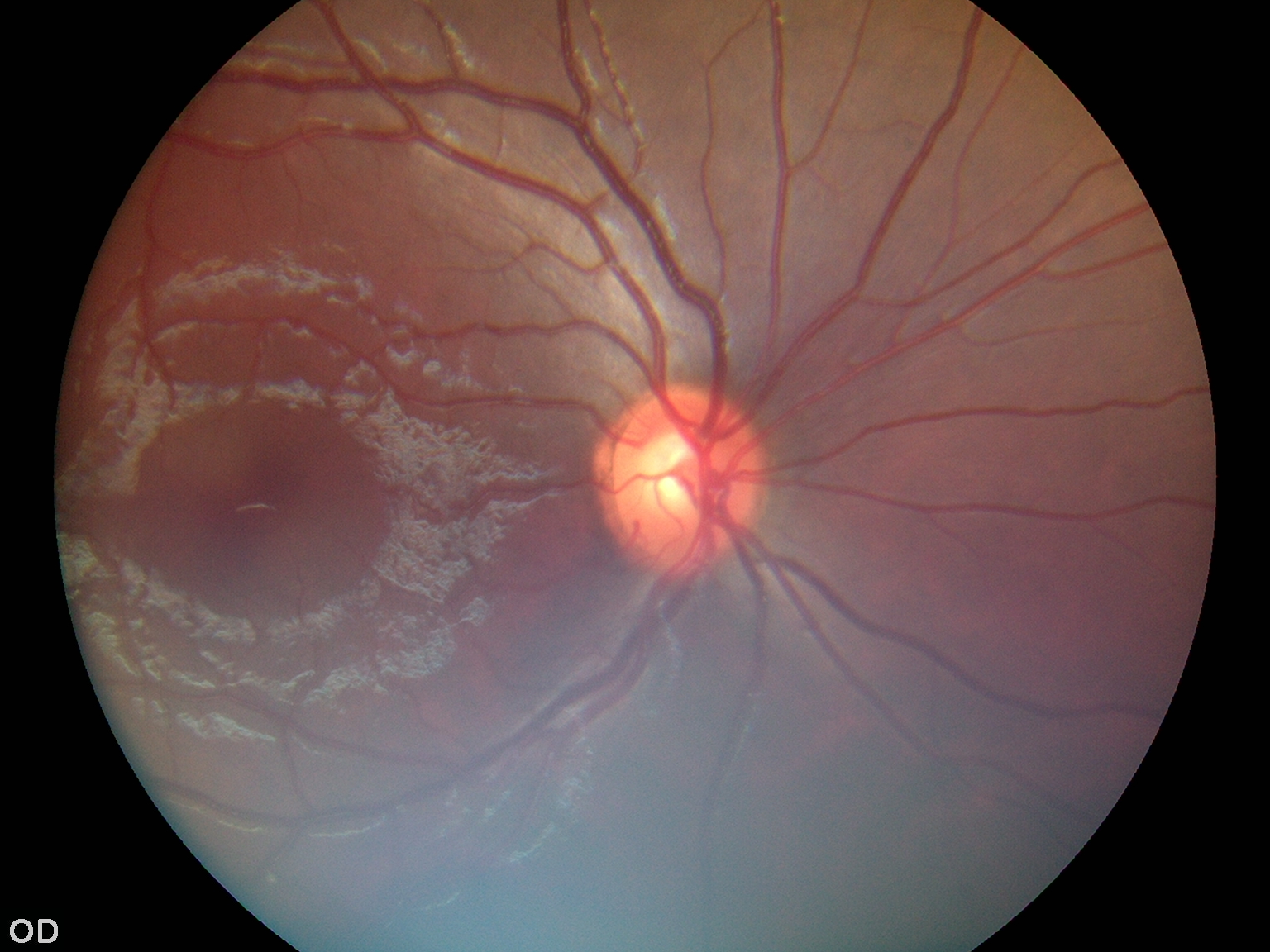
|
Retinal Fundus Image Dataset for Multi Disease Biomarker Analysis |
This dataset presents the initial release of 500 non-mydriatic color fundus photographs from a larger ongoing collection aimed at investigating retinal biomarkers for systemic diseases, with a primary focus on cardiovascular disease and stroke. The images were collected at the Department of Ophthalmology, Amrita Institute of Medical Sciences, Kochi, India, between January 2024 and the present. All photographs were acquired using a handheld Zeiss Visuscout 100 fundus camera under standard, non-dilated clinical imaging conditions. Each participant underwent bilateral imaging, capturing both macula centered and optic disc centered views in accordance with clinical protocols. All images have been fully anonymized to remove any personally identifiable information. The images are stored in JPG format, with an average file size of 1.03 MB per image and a standardized resolution suitable for computational analysis. To support quantitative and automated research, retinal vessel segmentation and the identification of disease related patterns have been performed using advanced deep learning architectures. The current release serves as a representative subset of the full dataset, which will comprise over 10,000 images in future phases. Associated clinical metadata, including basic demographic details, relevant medical history, and laboratory results, are available to enhance the dataset’s research value. Given the clinical sensitivity of the data, access is restricted and will be provided upon request to qualified researchers for non-commercial, academic, or clinically relevant purposes. This dataset is intended for reuse in machine learning, image analysis, and algorithm development, with the goal of advancing understanding of the links between retinal health and systemic diseases in diverse populations.
|
OPTHS_1000000029
(Request Access)
Views: 1
Images: 500
Downloads: 0
Data size: N/A
|
Indian Retinal Fundus Image Repository for Multi Disease Biomarker Analysis |
This dataset marks the initial release of 500 non-mydriatic retinal fundus photographs, forming part of a larger, ongoing effort to explore retinal biomarkers associated with systemic diseases, particularly cardiovascular disease and stroke. The images were collected at the Department of Ophthalmology, Amrita Institute of Medical Sciences in Kochi, India, from January 2024 onward. Using a handheld Zeiss Visuscout 100 fundus camera, bilateral retinal imaging was performed under standard clinical conditions without pupil dilation. Each subject contributed macula-centered and optic disc-centered views, following established ophthalmic protocols. All images have been anonymized to ensure patient privacy and are stored in JPG format, averaging 1.03 MB per image. The resolution is standardized to support computational analysis. To facilitate automated research, advanced deep learning techniques have been applied for retinal vessel segmentation and pattern recognition related to disease markers. This release represents a curated subset of a larger dataset that will eventually include over 10,000 images. Accompanying clinical metadata such as demographic information, medical history, and lab results enhances the dataset’s utility for research. Due to the sensitive nature of the data, access is restricted to qualified researchers and is granted upon request for non-commercial, academic, or clinically relevant use. The dataset is designed to support machine learning, image analysis, and algorithm development, with the overarching goal of improving understanding of the relationship between retinal health and systemic diseases across diverse populations. It offers a valuable resource for advancing precision diagnostics and predictive modelling in ophthalmology and systemic disease research.
|
|
Amrita Vishwa Vidyapeetham, Coimbatore |
Managed Access
|
June 3, 2026 |
IBIAP_1000000026

|
Dataset of Centella Asiatica leaves for quality assessment and machine learning applications |
Centella asiatica is a significant medicinal herb extensively used in traditional oriental medicine and gaining global popularity. The primary constituents of Centella asiatica leaves are triterpenoid saponins, which are predominantly believed to be responsible for its therapeutic properties. Ensuring the use of high-quality leaves in herbal medicine preparation is crucial across all medicinal practices. To address this quality control issue using machine learning applications, we have developed an image dataset of Centella asiatica leaves. The images were captured using Samsung Galaxy M21 mobile phones and depict the leaves in “Dried,” “Healthy,” and “Unhealthy” states. These states are further divided into “Single” and “Multiple” leaves categories, with “Single” leaves being further classified into “Front” and “Back” views to facilitate a comprehensive study. The images were pre-processed and standardized to 1024 × 768 dimensions, resulting in a dataset comprising a total of 9094 images. This dataset is instrumental in the development and evaluation of image recognition algorithms, serving as a foundational resource for computer vision research. Moreover, it provides a valuable platform for testing and validating algorithms in areas such as image categorization and object detection. For researchers exploring the medicinal potential of Centella asiatica in traditional medicine, this dataset offers critical information on the plantʼs health, thereby advancing research in herbal medicine and ethnopharmacology.
|
PPS_1000000031
(Download Images)
(Print Records)
Views: 1
Images: 9094
Downloads: 0
Data size: 1.4GB
|
Image Dataset of Centella Asiatica for quality assessment and machine learning applications |
This Centella asiatica dataset is structured into three main folders representing various states of leaf health: Dried (2996 images), which are organized into Single (2018 images) and Multiple (985 images) subfolders. Unlike the healthy and unhealthy categories, the Single images of dried leaves are not divided into front and back views due to the shriveled nature of dried leaves, which obscures these distinctions., Healthy (3048 images) and is further divided into Single (2044 images) and Multiple (1004 images) subfolders. The Single subfolder contains images of individual leaves and is further categorized by leaf orientation into Front (1014 images) and Back (1030 images) views , and Unhealthy (3050 images), this folder is similarly organized into Single (2024 images) and Multiple (1016 images) subfolders. The Single images are further divided into Front (1023 images) and Back (1011 images) views. This organization enables machine learning models to classify leaves based on health status, facilitating plant quality assessment and real-time health monitoring in agricultural systems. Each main folder is further divided to capture detailed leaf views, thereby supporting complex image recognition and classification tasks. This comprehensive dataset not only facilitates Centella asiatica identification but also supports a range of machine learning applications in plant health assessment, quality control, and agricultural decision-making. It is poised to aid researchers in developing machine learning models for the detection of adulteration in herbal products, a critical need in ensuring the authenticity of herbal medicine. Additionally, the dataset's structured diversity in health states and orientations makes it an invaluable resource for computer vision researchers focused on fine-grained plant image classification and the development of robust, application-ready models. By offering categorized, high-quality images, this dataset provides a foundation for advancing both applied research and the broader fields of ethnobotany and agricultural informatics.
|
https://doi.org/10.1016/j.dib.2024.111150 |
Vishwakarma University, Pune |
Open Access
|
Nov. 14, 2025 |
IBIAP_1000000027

|
PCSeg: Color model driven probabilistic multiphase level set based tool for plasma cell segmentation in multiple myeloma |
Plasma cell segmentation is the first stage of a computer assisted automated diagnostic tool for multiple myeloma (MM). Owing to large variability in biological cell types, a method for one cell type cannot be applied directly on the other cell types. In this study, PCSeg Tool for plasma cell segmentation from microscopic medical images has presented. These images were captured from bone marrow aspirate slides of patients with multiple myeloma as per the standard guidelines. PCSeg has a robust pipeline consisting of a pre-processing step, the proposed modified multiphase level set method followed by post-processing steps including the watershed and circular Hough transform to segment clusters of cells of interest and to remove unwanted cells. Our modified level set method utilizes prior information about the probability densities of regions of interest (ROIs) in the color spaces and provides a solution to the minimal-partition problem to segment ROIs in one of the level sets of a two-phase level set formulation. PCSeg tool is tested on a number of microscopic images and provides good segmentation results on single cells as well as efficient segmentation of plasma cell clusters.
|
HISTOS_1000000032
(Download Images)
(Print Records)
Views: 1
Images: 85
Downloads: 0
Data size: 805MB
|
MiMM_SBILab Dataset: Microscopic Images of Multiple Myeloma |
This dataset is a collection of 85 microscopic images of Multiple Meyloma subjects with a number of plasma cells marked by the expert pathologist. We proposed a method of color-driven multiphase levelset for automating cell segmentation in Multiple Myeloma that has been published in PLOS One Journal. The users of this dataset would be required to cite that paper. This is an effort towards building an automated pipeline for cancer detection in Multiple Myeloma. Interested researchers can propose deep learning based or advanced machine learning based solutions for plasma cell segmentation using this dataset.
|
https://doi.org/10.1371/journal.pone.0207908 |
Indraprastha Institute of Information Technology, Delhi |
Open Access
|
Nov. 14, 2025 |