Deposition Date
2023-06-05
Release Date
2023-11-29
Last Version Date
2025-07-02
Entry Detail
PDB ID:
8JMO
Keywords:
Title:
Structure of a leaf-branch compost cutinase, ICCG in complex with 4-((4-Hydroxybutoxy)carbonyl)benzoic acid
Biological Source:
Source Organism:
unidentified prokaryotic organism (Taxon ID: 2725)
Host Organism:
Method Details:
Experimental Method:
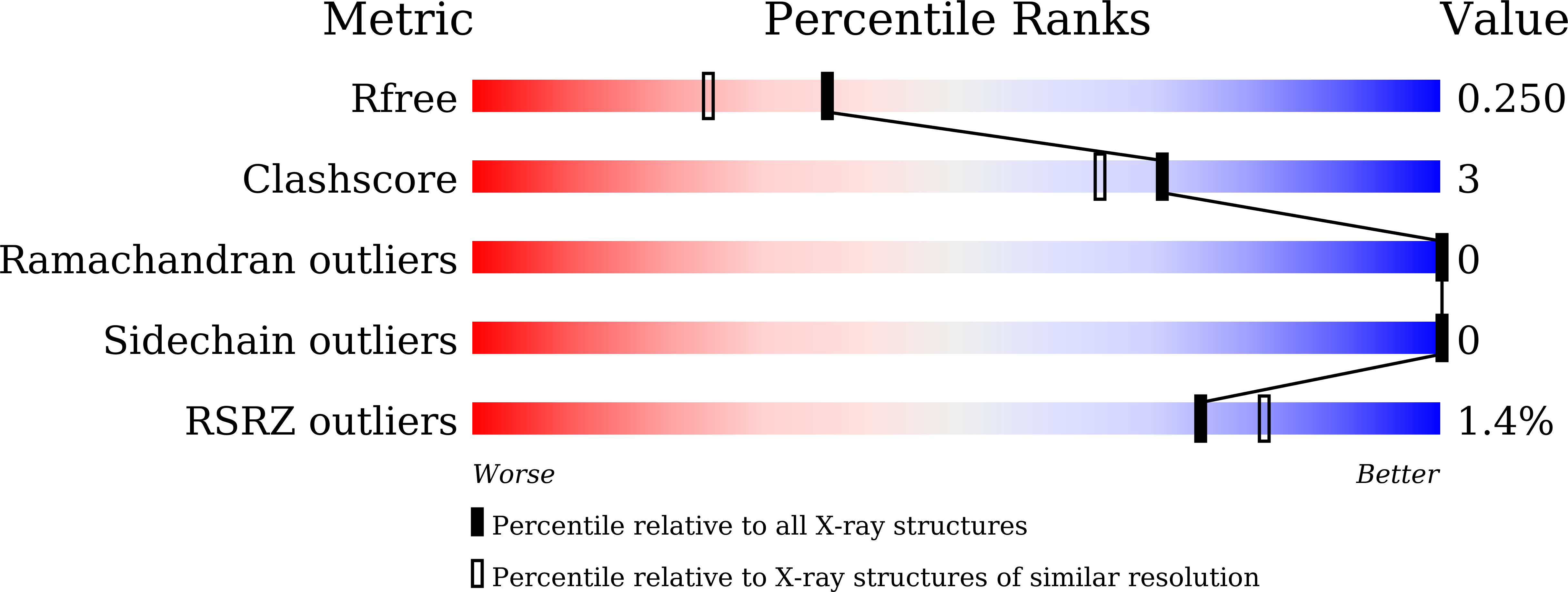
Resolution:
1.95 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 21