Deposition Date
2016-09-09
Release Date
2017-09-27
Last Version Date
2024-11-13
Entry Detail
PDB ID:
5GW8
Keywords:
Title:
Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
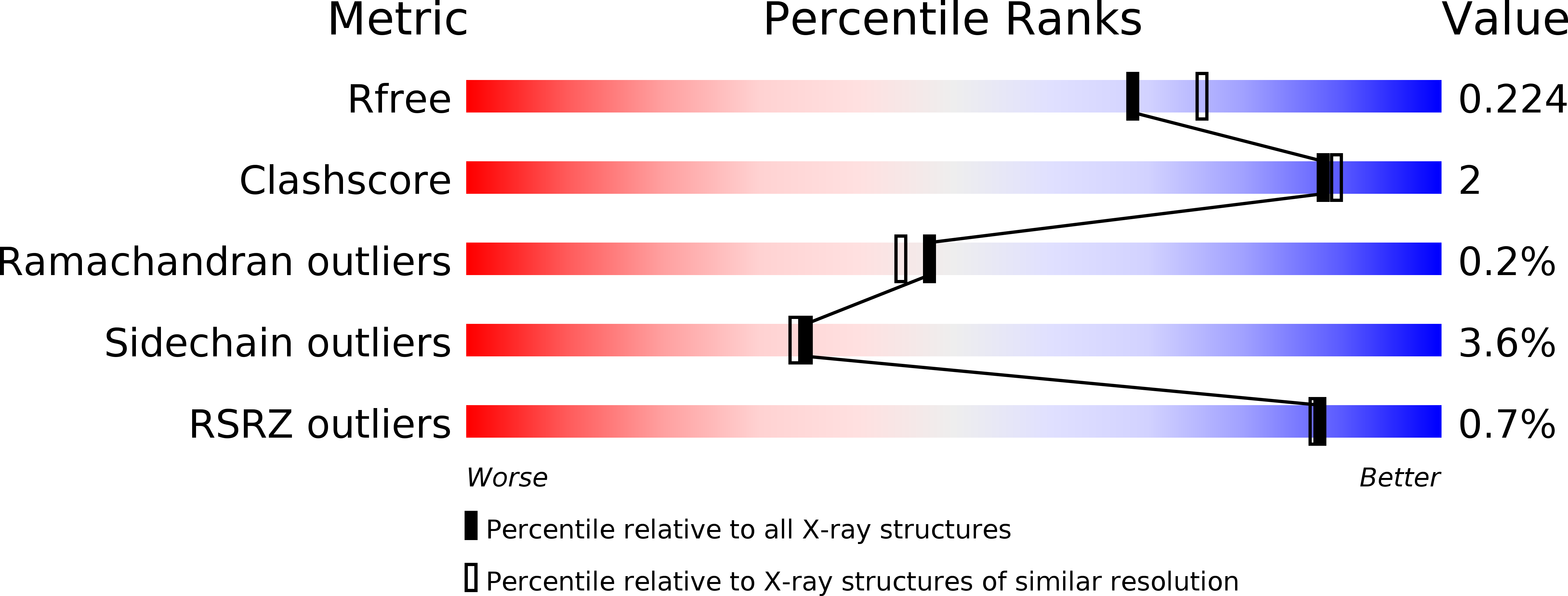
Resolution:
2.00 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 21 21 21