Search Count: 19
 |
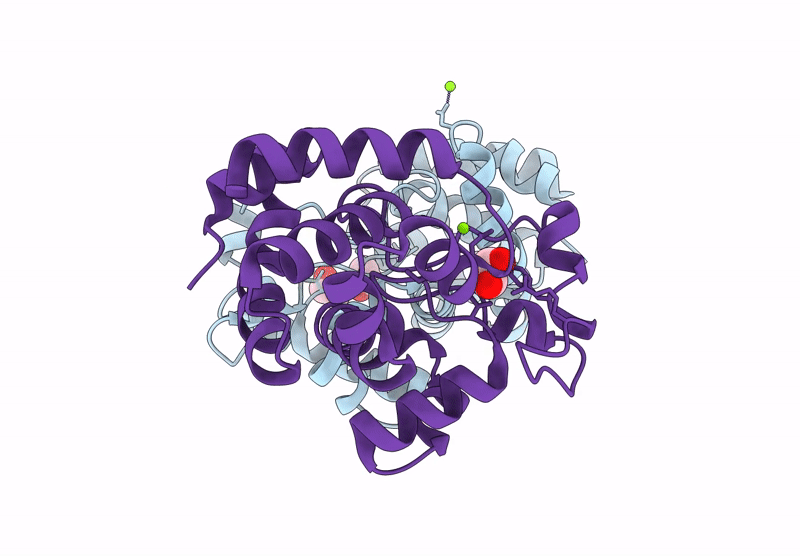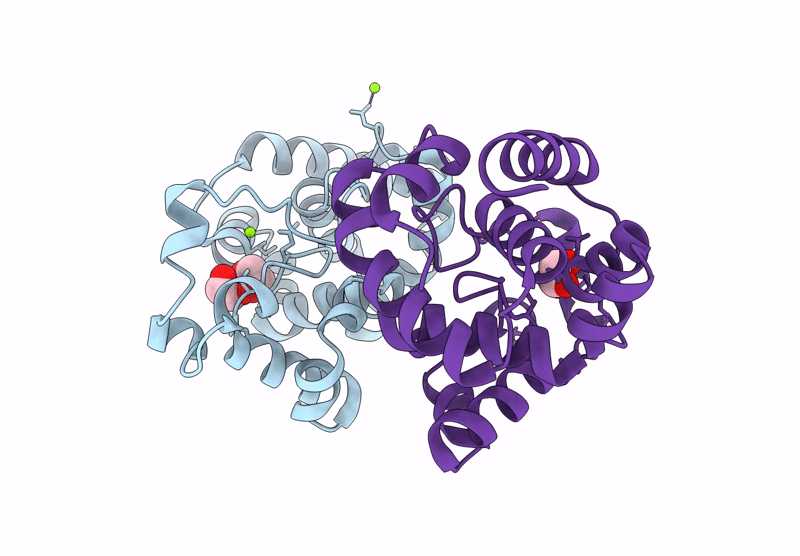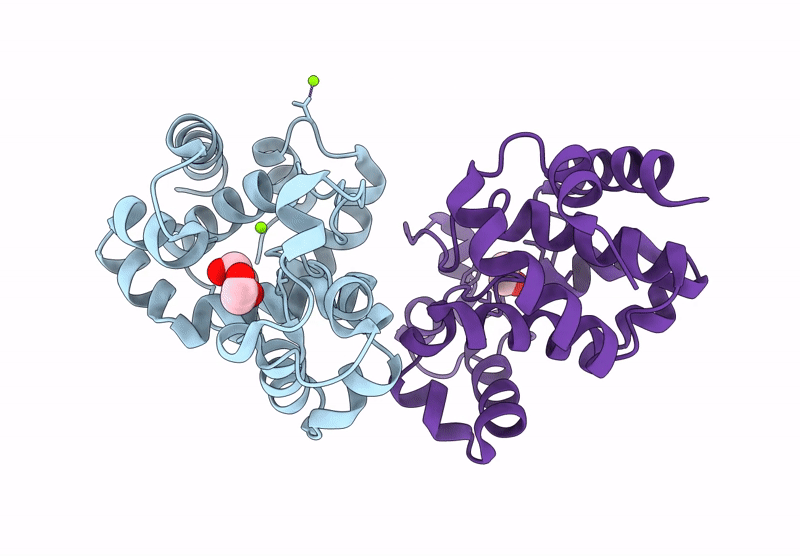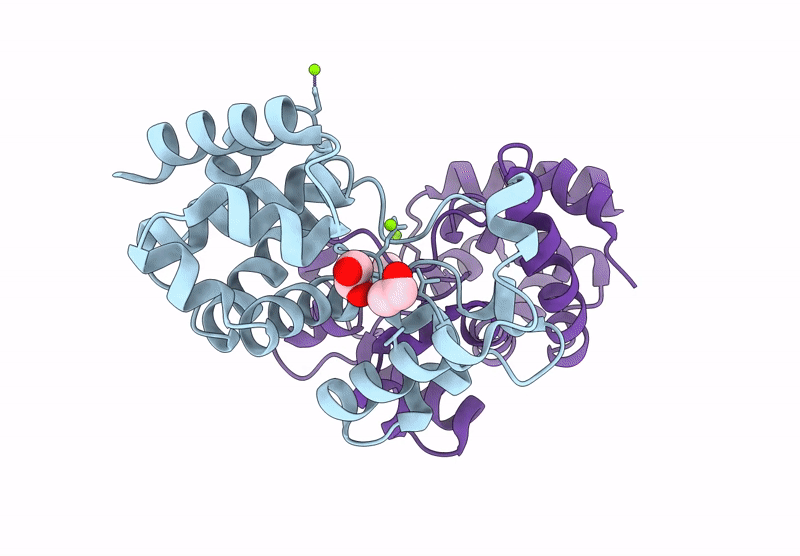
Crystal Structure Of A Putative Phage Endolysin Identified From A Metagenomic Survey Of Prosser, Washington Soil (Pwe1)
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: SO4, BR |
 |
Crystal Structure Of A Putative Phage Endolysin Identified From A Metagenomic Survey Of Prosser, Washington Soil (Pwe2)
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: MG, EDO, CL |
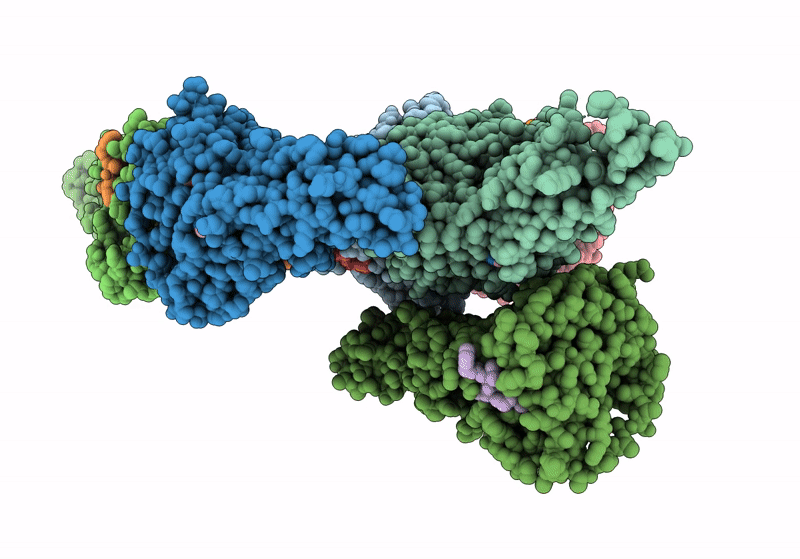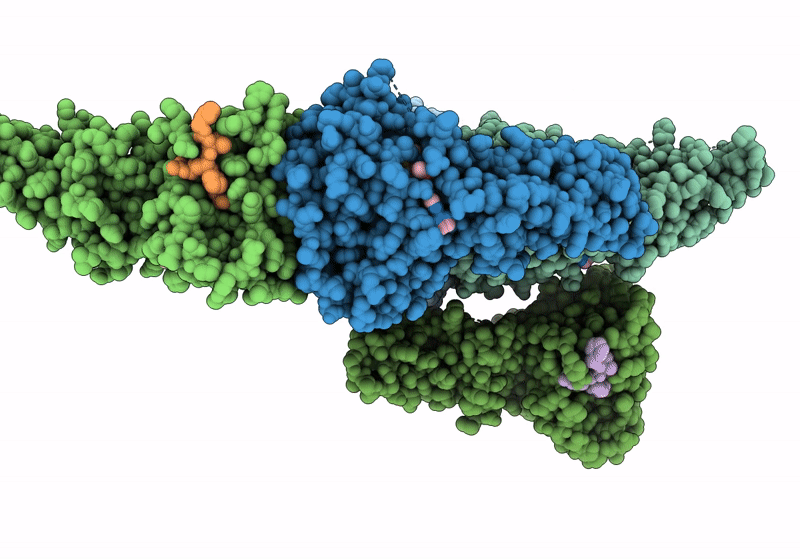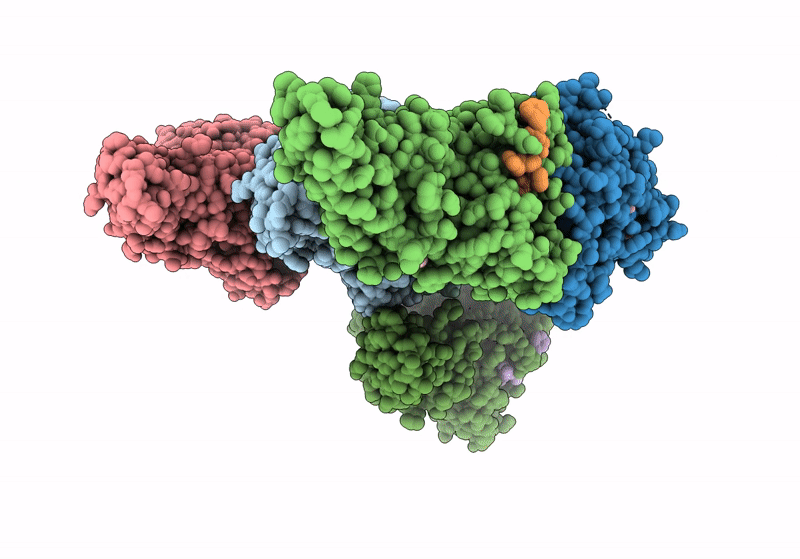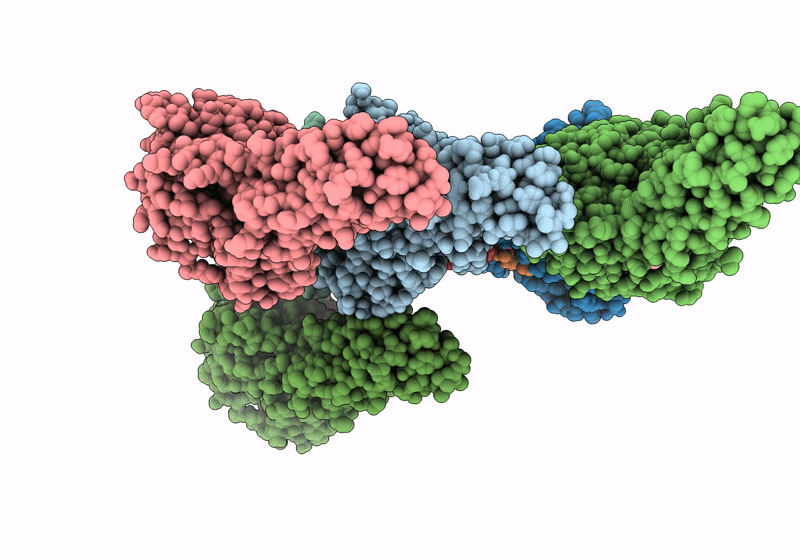 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR Ligands: A1AAB |
 |
Organism: Organismal metagenomes
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: GOL, CL |
 |
Organism: Prolixibacter bellariivorans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-04-13 Classification: HYDROLASE |
 |
Crystal Structure Of Carbohydrate Esterase Pbeacxe, In Complex With Meglcpa-Xylp
Organism: Prolixibacter bellariivorans
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2022-04-13 Classification: HYDROLASE |
 |
Crystal Structure Of Carbohydrate Esterase Pbeacxe, In Complex With Acetate
Organism: Prolixibacter bellariivorans
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2022-04-13 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Chryseobacterium sp. yr480
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-04-13 Classification: HYDROLASE Ligands: CL |
 |
Organism: Flavobacterium johnsoniae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-04-13 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-03-02 Classification: SIGNALING PROTEIN, Transferase Ligands: 9ID, SO4 |
 |
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: MPD, BTB |
 |
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: ACT, MPD, BTB, AHR |
 |
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: 1PE, PG4, CL, PGE |
 |
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: AHR, MPD, ACT, BTB |
 |
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-02-10 Classification: HYDROLASE |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: AES |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-12-16 Classification: HYDROLASE Ligands: SO4 |
 |
Human Carbonic Anhydrases Ii In Complex With A Acetazolamide Derivative Comprising One Hydrophobic And One Hydrophilic Tail Moiety
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2015-01-28 Classification: Lyase/Lyase inhibitor Ligands: ZN, FMT, 3T7, GOL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-30 Classification: CELL CYCLE Ligands: CIT, K |

