Search Count: 17
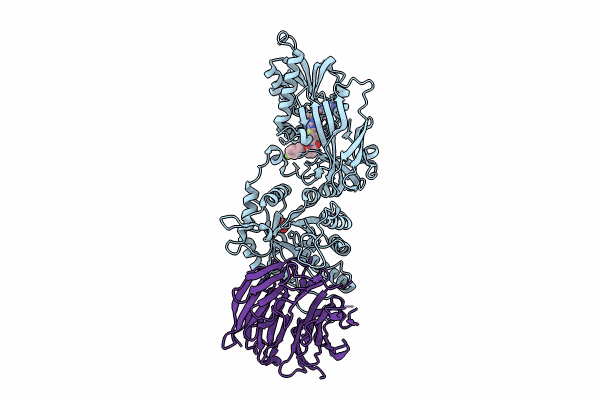 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-08-14 Classification: TRANSFERASE Ligands: A1H8A, MTA, SO4 |
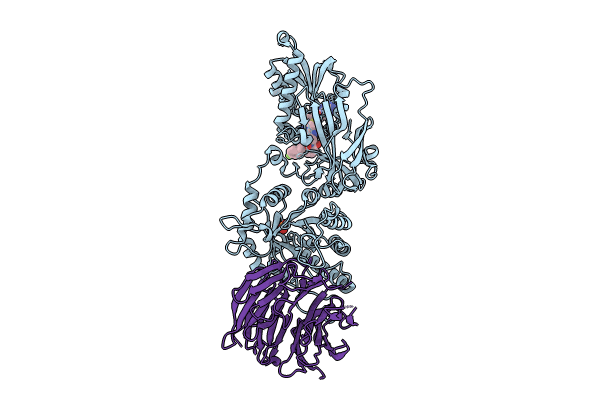 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-08-14 Classification: TRANSFERASE Ligands: A1H76, MTA, SO4 |
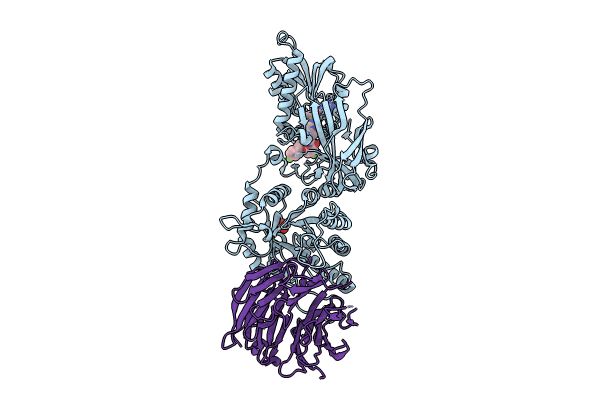 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-14 Classification: TRANSFERASE Ligands: A1H8B, MTA, SO4 |
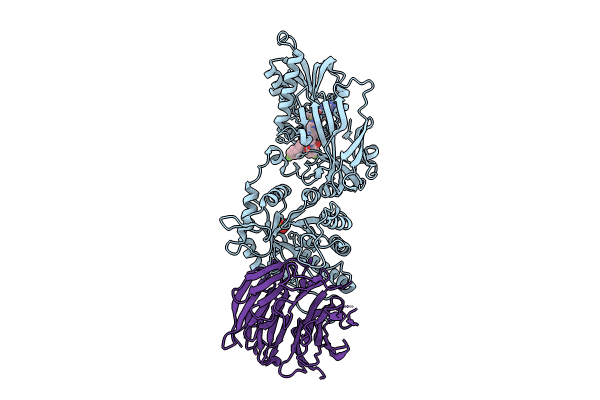 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-08-14 Classification: TRANSFERASE Ligands: A1H73, MTA, SO4 |
 |
Structure Of Mycobacterium Smegmatis Alpha-Maltose-1-Phosphate Synthase Glgm
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-22 Classification: SUGAR BINDING PROTEIN |
 |
Glge Isoform 1 From Streptomyces Coelicolor E423A Mutant Soaked In Maltoheptaose
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-17 Classification: TRANSFERASE |
 |
Glge Isoform 1 From Streptomyces Coelicolor E423A Mutant Soaked In Maltooctaose
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-17 Classification: TRANSFERASE |
 |
Glge Isoform 1 From Streptomyces Coelicolor D394A Mutant Co-Crystallised With Maltodextrin
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-08-17 Classification: HYDROLASE Ligands: EDO, CIT |
 |
Glge Isoform 1 From Streptomyces Coelicolor D394A Mutant With Maltose- 1-Phosphate Bound
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-05-21 Classification: HYDROLASE |
 |
Glge Isoform 1 From Streptomyces Coelicolor E423A Mutant With 2-Deoxy- 2-Fluoro-Beta-Maltosyl Modification
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-05-21 Classification: TRANSFERASE |
 |
Glge Isoform 1 From Streptomyces Coelicolor E423A Mutant With Maltose Bound
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-05-21 Classification: HYDROLASE |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-09-14 Classification: HYDROLASE |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-14 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-09-14 Classification: HYDROLASE |
 |
Glge Isoform 1 From Streptomyces Coelicolor With Alpha-Cyclodextrin And Maltose Bound
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2011-09-14 Classification: HYDROLASE |
 |
Glge Isoform 1 From Streptomyces Coelicolor With Beta-Cyclodextrin And Maltose Bound
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-09-14 Classification: HYDROLASE |
 |
Dnab Binding Domain Of Dnag (P16) From Bacillus Stearothermophilus (Residues 452-597)
Organism: Geobacillus stearothermophilus
Method: SOLUTION NMR Release Date: 2005-10-04 Classification: TRANSFERASE |

