Search Count: 8
 |
Organism: Halalkalibacterium halodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-08-31 Classification: DNA Ligands: EDO, CL, GOL, PEG, NA, ACT |
 |
Organism: Alkalihalobacillus halodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-08-31 Classification: DNA Ligands: GOL, EDO, ACT, NA, K |
 |
Organism: Halalkalibacterium halodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-08-31 Classification: DNA Ligands: EDO, GOL, NA |
 |
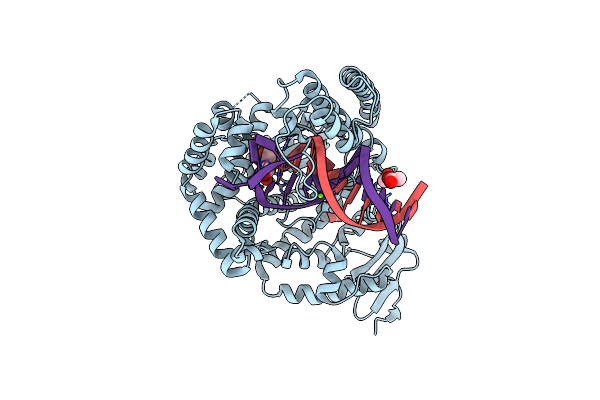
Crystal Structure Of Klentaq Mutant M747K In A Closed Ternary Complex With A Dg:Dctp Base Pair
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-29 Classification: TRANSFERASE Ligands: MG, DCP, GOL |
 |
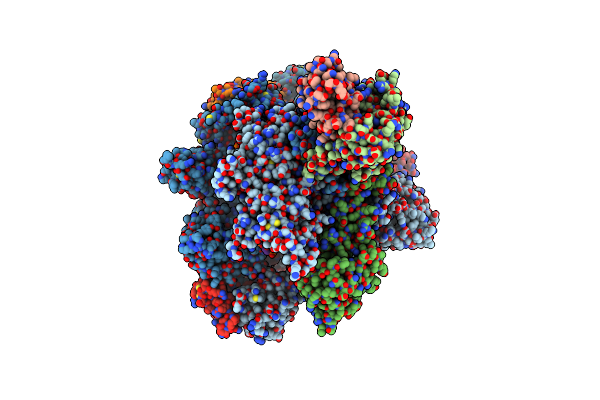
Crystal Structure Of Klentaq Mutant M747K In A Closed Ternary Complex With A O6-Meg:Benzitp Base Pair
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-11-29 Classification: TRANSFERASE Ligands: MN, NZI, ACT, MG |
 |
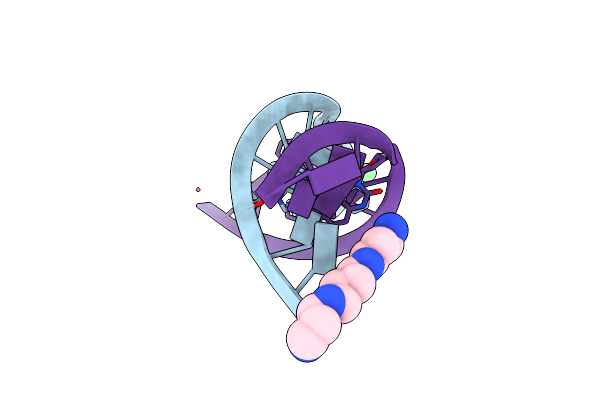
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2017-11-01 Classification: TRANSCRIPTION Ligands: ZN, MG, AHW |
 |
Structure Of O6-Benzyl-2'-Deoxyguanosine Opposite Perimidinone-Derived Synthetic Nucleoside In Dna Duplex
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-10 Classification: DNA Ligands: SPM, SR |
 |
Structure Of Perimidinone-Derived Synthetic Nucleoside Paired With Guanine In Dna Duplex
|

