Search Count: 55
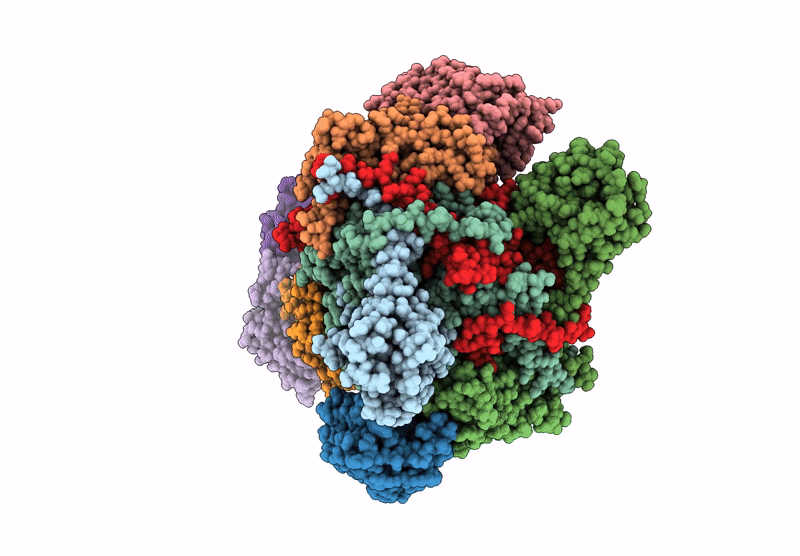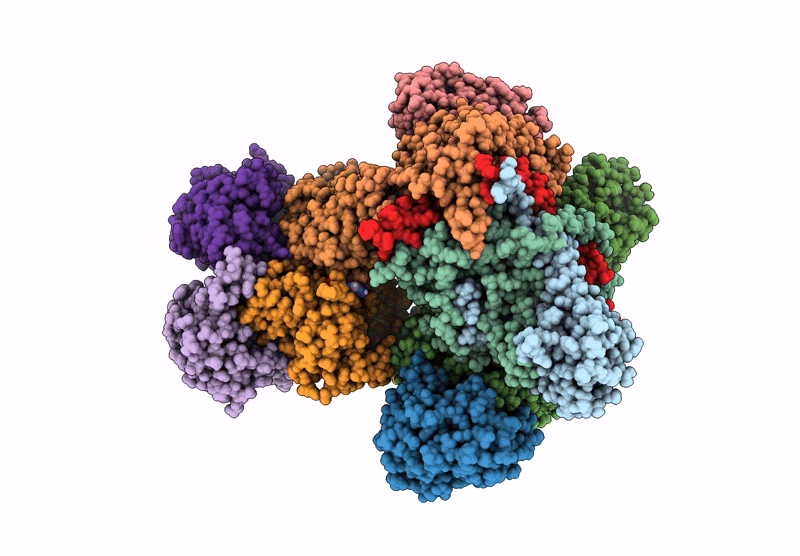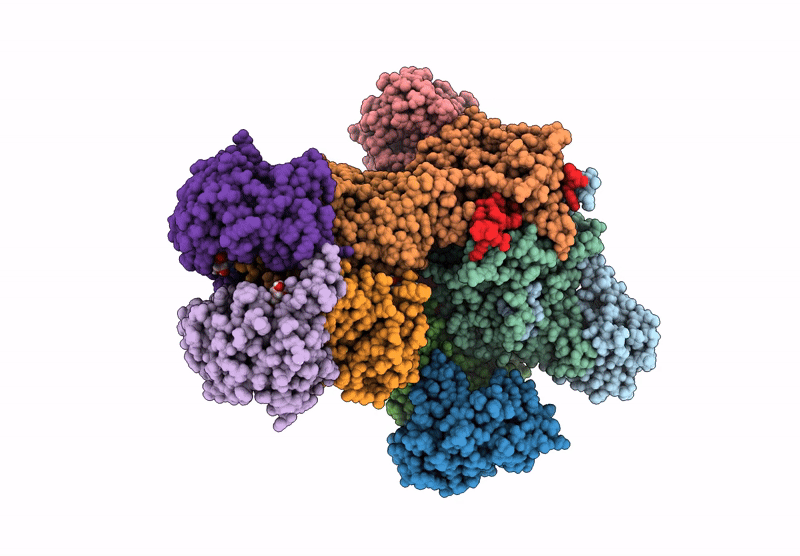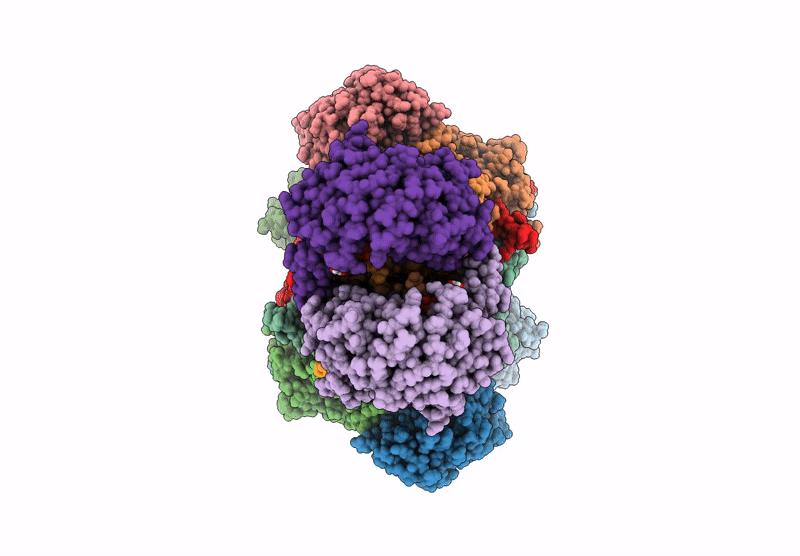 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: ZN, I9X, ADP, PO4, MG, ATP |
 |
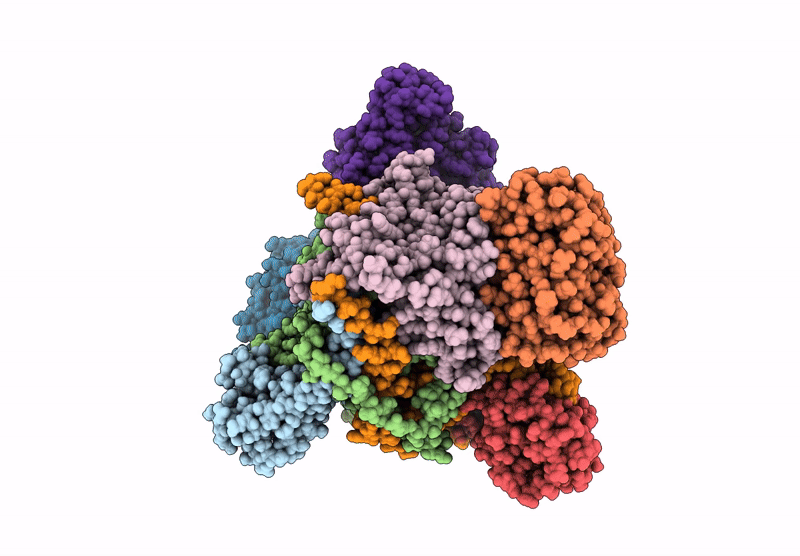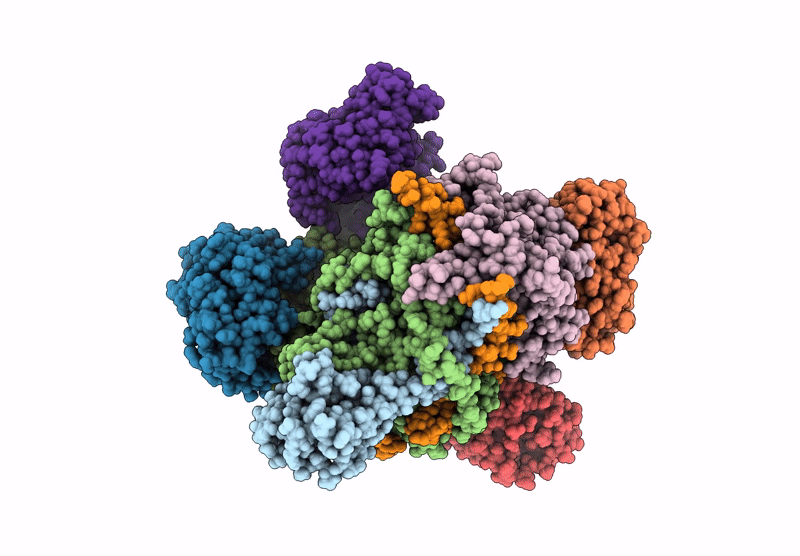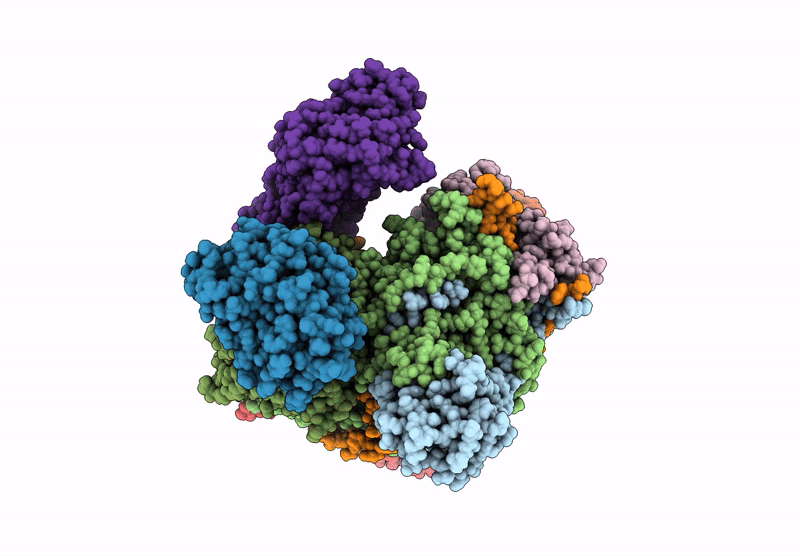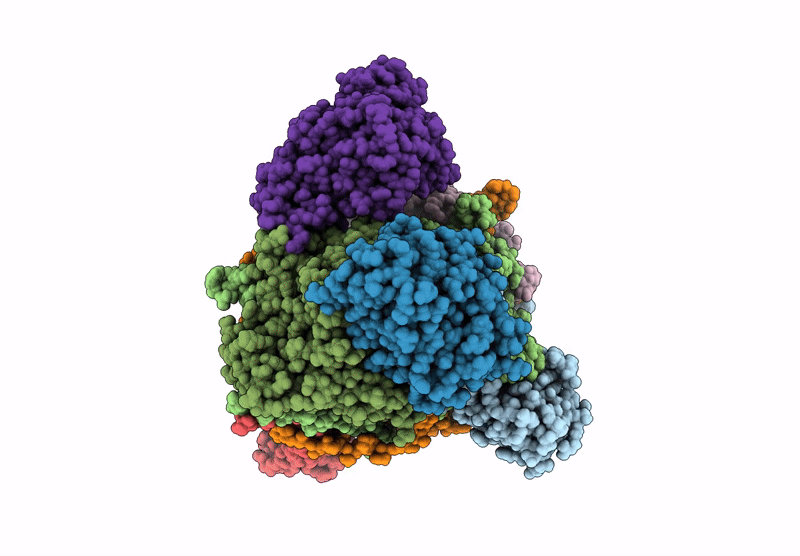
E. Coli C-P Lyase Bound To Phnk/Phnl Dual Abc Dimer With Amppnp And Phnk E171Q Mutation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: ZN, PO4, MG, ANP |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: TRANSFERASE Ligands: ZN, I9X |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: TRANSFERASE Ligands: ZN, I9X, ATP, MG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: TRANSFERASE Ligands: ZN, PO4 |
 |
Crystal Structure Of Desb30 Insulin Produced By Cell Free Protein Synthesis
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-06-16 Classification: HORMONE |
 |
Apo Form Of C-Type Lysozyme From The Upper Gastrointestinal Tract Of Opisthocomus Hoatzin
Organism: Opisthocomus hoazin
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-11-20 Classification: HYDROLASE Ligands: GOL, CL |
 |
Complex With Chitin Oligomer Of C-Type Lysozyme From The Upper Gastrointestinal Tract Of Opisthocomus Hoatzin
Organism: Opisthocomus hoazin
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2019-11-20 Classification: HYDROLASE Ligands: CL |
 |
Crystallographic Structure Of The Catalytic Domain Of Human Phenylalanine Hydroxylase (Hpah Cd) In Complex With Iron At 1.6 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-06-05 Classification: OXIDOREDUCTASE Ligands: FE |
 |
The Structure Of Full-Length Human Phenylalanine Hydroxylase In Complex With The Cofactor And Negative Regulator Tetrahydrobiopterin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2019-06-05 Classification: OXIDOREDUCTASE Ligands: H4B |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-12-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-10-24 Classification: TOXIN Ligands: IMD, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-10-25 Classification: Hydrolase/Inhibitor Ligands: SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase (Fr Conformation)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, PEG, SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase Exposed To Visible Light (30 Min)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, PEG, SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase Exposed To Visible Light (60 Min)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, PEG, SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase Exposed To Visible Light (120 Min)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, PEG, SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase Exposed To Visible Light (180 Min)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, PEG, SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase Exposed To Visible Light (240 Min)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, PEG, SO4 |
 |
Crystal Structure Of Lactococcus Lactis Thioredoxin Reductase (Fo Conformation)
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |

