Search Count: 8
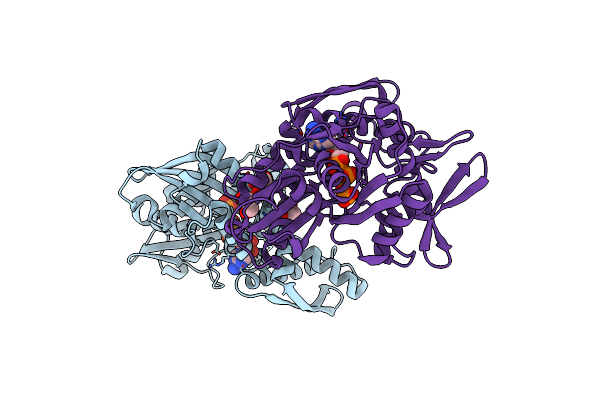 |
Disrupted Hydrogen Bond Network Impairs Atpase Activity In An Hsc70 Cysteine Mutant
Organism: homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-01-17 Classification: CHAPERONE Ligands: MG, PO4, ADP, GOL |
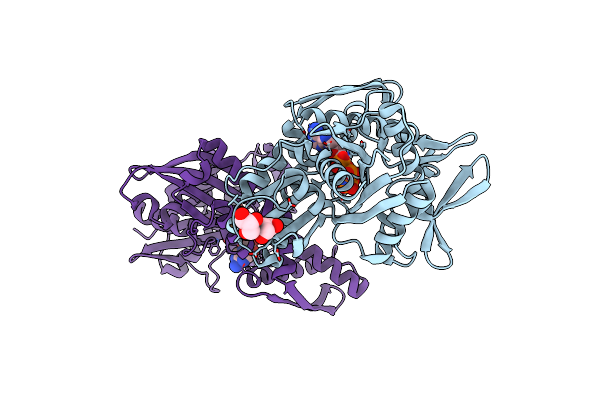 |
Disrupted Hydrogen Bond Network Impairs Atpase Activity In An Hsc70 Cysteine Mutant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-17 Classification: CHAPERONE Ligands: ANP, MG, GOL |
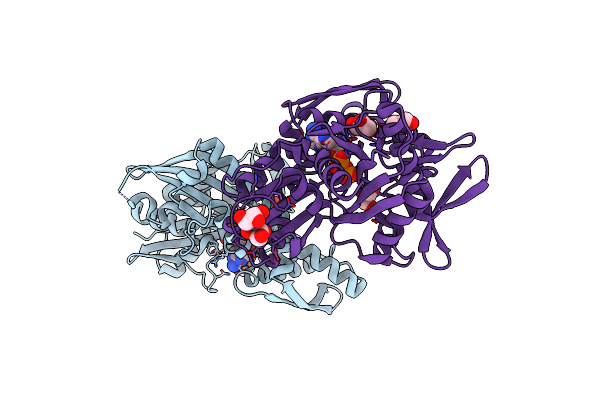 |
Disrupted Hydrogen Bond Network Impairs Atpase Activity In An Hsc70 Cysteine Mutant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-01-17 Classification: CHAPERONE Ligands: ADP, MG, GOL |
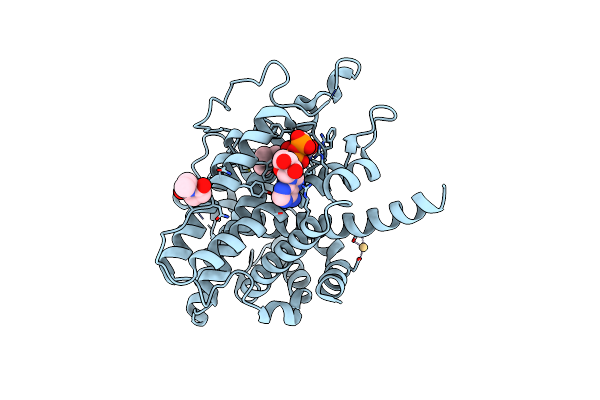 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-11-03 Classification: OXIDOREDUCTASE Ligands: NEN, FAD, CD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2010-11-03 Classification: OXIDOREDUCTASE Ligands: NEN, FAD, CD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-04 Classification: ISOMERASE Ligands: ACT, EDO |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2001-12-28 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-12-28 Classification: OXIDOREDUCTASE Ligands: FAD |

