Search Count: 48
 |
Re-Engineered Peroxygenase Variant Of 2-Deoxy-D-Ribose-5-Phosphate Aldolase In Substrate-Free State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-03-12 Classification: LYASE Ligands: GOL |
 |
Re-Engineered Peroxygenase Variant Of 2-Deoxy-D-Ribose-5-Phosphate Aldolase, Schiff-Base Complex With 4-Chloro-Cinnamaldehyde
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-03-12 Classification: LYASE Ligands: A1ICA, GOL |
 |
Re-Engineered Peroxygenase Variant Of 2-Deoxy-D-Ribose-5-Phosphate Aldolase, Schiff-Base Complex With 4-Nitro-Cinnamaldehyde
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-03-12 Classification: LYASE Ligands: A1IB9, GOL |
 |
Organism: Bacillus tequilensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, CL, MG |
 |
Organism: Pyrenophora teres f. teres 0-1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Pyrenophora teres f. teres 0-1
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN Ligands: P1T |
 |
Structure Of A Fused 4-Ot Variant Engineered For Asymmetric Michael Addition Reactions
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-01-26 Classification: LYASE Ligands: GOL, CL |
 |
Re-Engineered 2-Deoxy-D-Ribose-5-Phosphate Aldolase Catalysing Asymmetric Michael Addition Reactions In Substrate-Free State
Organism: Escherichia coli (strain atcc 8739 / dsm 1576 / nbrc 3972 / ncimb 8545 / wdcm 00012 / crooks)
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2021-10-27 Classification: LYASE |
 |
Re-Engineered 2-Deoxy-D-Ribose-5-Phosphate Aldolase Catalysing Asymmetric Michael Addition Reactions, Schiff Base Complex With Cinnamaldehyde
Organism: Escherichia coli 909945-2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-27 Classification: LYASE Ligands: 9Y6, GOL |
 |
Glutamate Transporter Homologue Glttk In Complex With A Photo Cage Compound
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: QJW, PEG, PGE |
 |
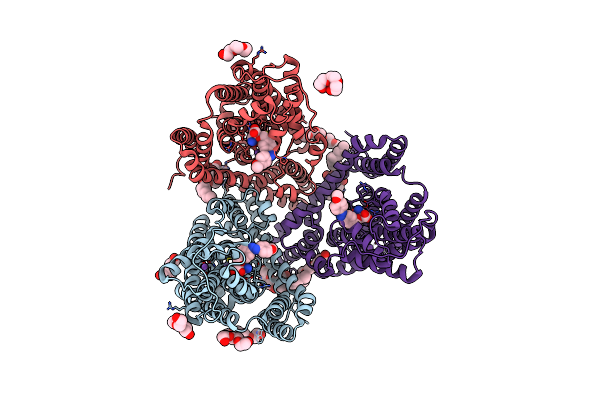
The Structure Of Glutamate Transporter Homologue Glttk In Complex With The Photo Switchable Compound (Cis)
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: QM5, DMU, PG4, PGE, NA, PEG, 1PE |
 |
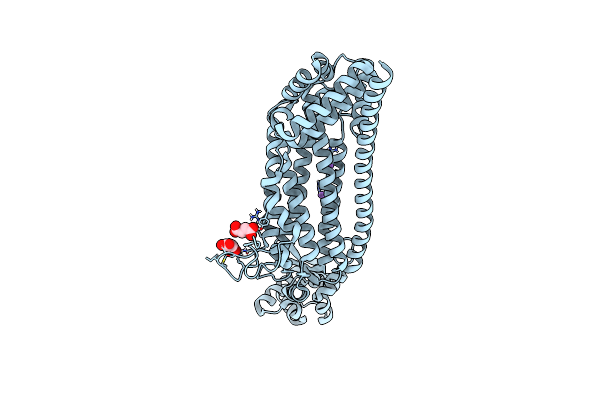
The Structure Of Glutamate Transporter Homologue Glttk In Complex With The Photo Switchable Compound (Trans)
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-01-27 Classification: TRANSPORT PROTEIN Ligands: QM5, DMU, PG4, PGE, NA, PEG, 1PE |
 |
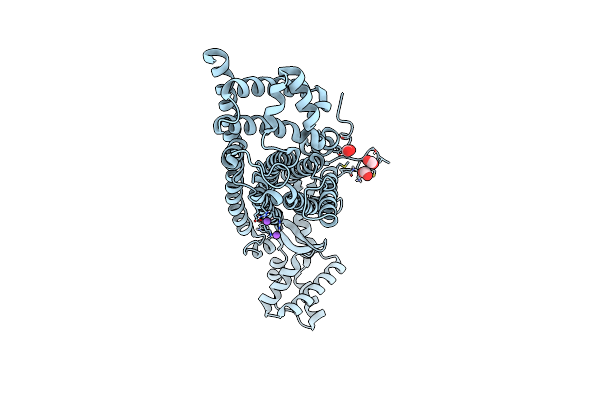
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-10-30 Classification: LYASE Ligands: FUM, NA |
 |
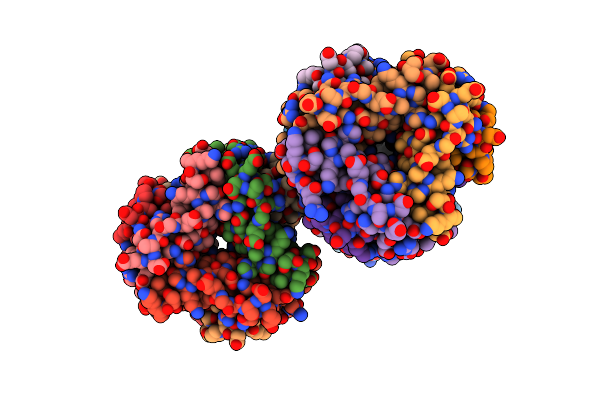
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-10-30 Classification: LYASE Ligands: FMT, GOL, NA |
 |
Crystal Structure Of 4-Oxalocrotonate Tautomerase Triple Mutant L8Y/M45Y/F50A
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-06 Classification: ISOMERASE Ligands: PO4, GOL |
 |
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-05-16 Classification: LYASE |
 |
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-16 Classification: LYASE Ligands: FMT, NA |
 |
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-05-16 Classification: LYASE Ligands: FUM, PEG |
 |
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-05-16 Classification: LYASE Ligands: SIN, PEG |
 |
Organism: Chelativorans sp. (strain bnc1)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-05-16 Classification: LYASE Ligands: EKQ |

