Search Count: 26
 |
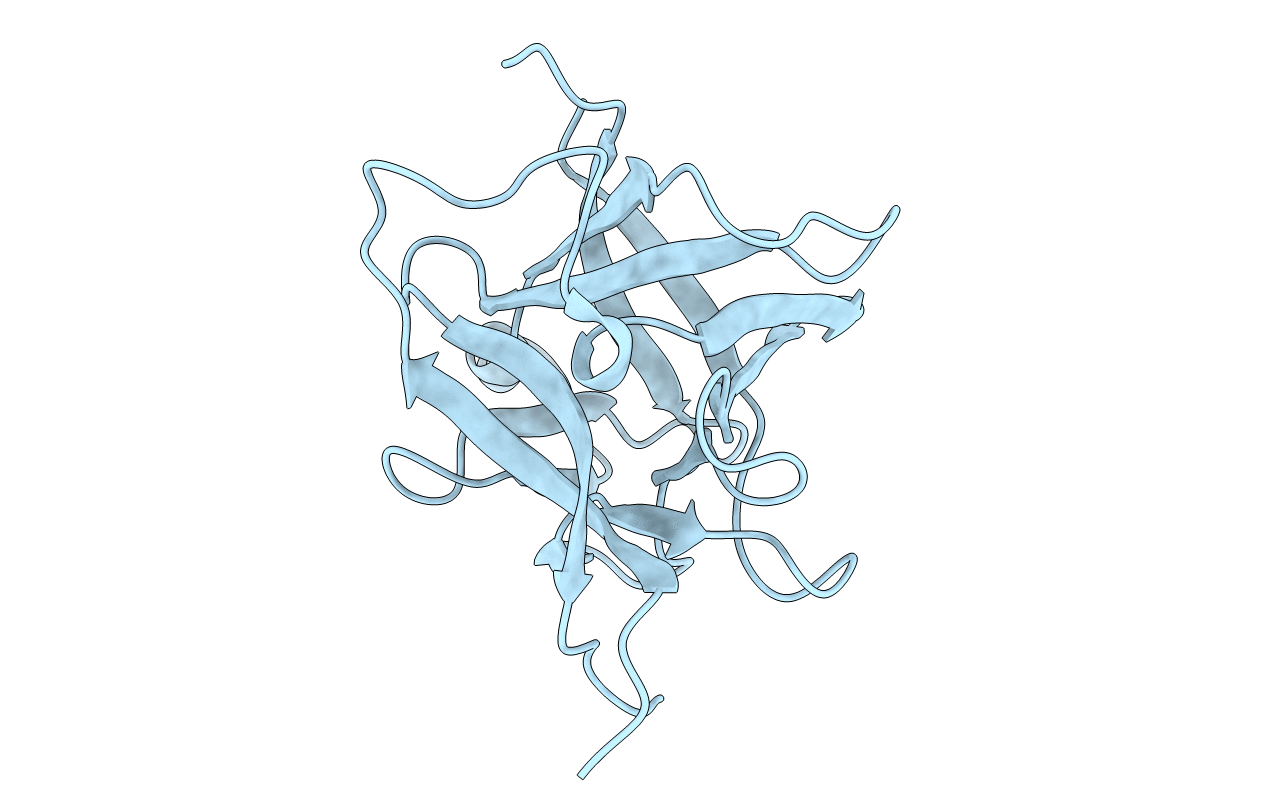
Structure Of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-02-16 Classification: CYTOKINE Ligands: TB, ACT, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-01-19 Classification: CYTOKINE |
 |
Crystal Structure Of Apo Form Of D,D-Heptose 1.7-Bisphosphate Phosphatase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: ZN, ACY |
 |
Crystal Structure Of D,D-Heptose 1.7-Bisphosphate Phosphatase From E. Coli Complexed With Magnesium And Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: MG, ZN, PO4 |
 |
Crystal Structure Of D,D-Heptose 1.7-Bisphosphate Phosphatase From E. Coli Complexed With D-Glycero-D-Manno-Heptose 1 ,7-Bisphosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: MG, ZN, NA, GMB |
 |
Crystal Structure Of D,D-Heptose 1.7-Bisphosphate Phosphatase From B. Bronchiseptica Complexed With Magnesium And Phosphate
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: MG, ZN, PO4, FMT, FX1, SAR |
 |
Analysis Of The Structural Determinants Underlying Discrimination Between Substrate And Solvent In Beta-Phosphoglucomutase Catalysis
Organism: Lactococcus lactis subsp. lactis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-06-09 Classification: ISOMERASE Ligands: MG |
 |
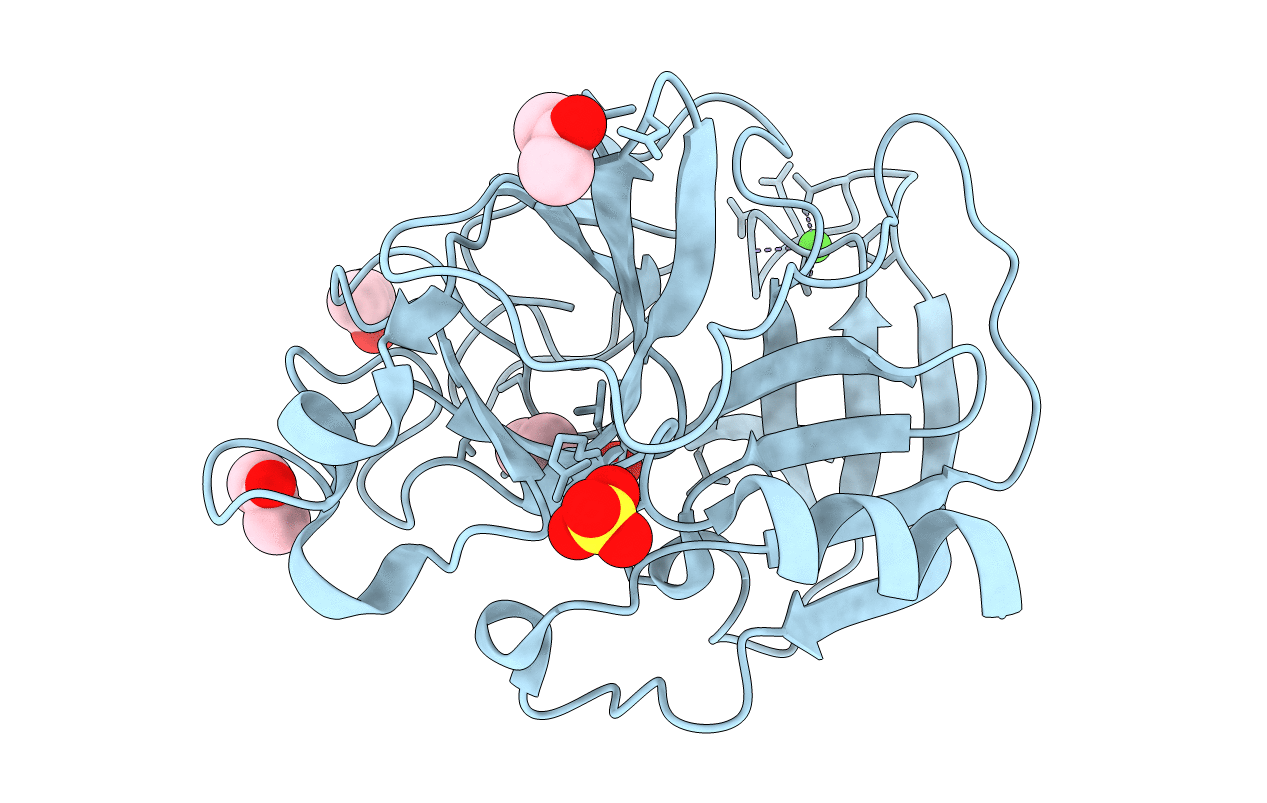
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-09-04 Classification: PROTEIN BINDING Ligands: MG, EPE |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, ACN |
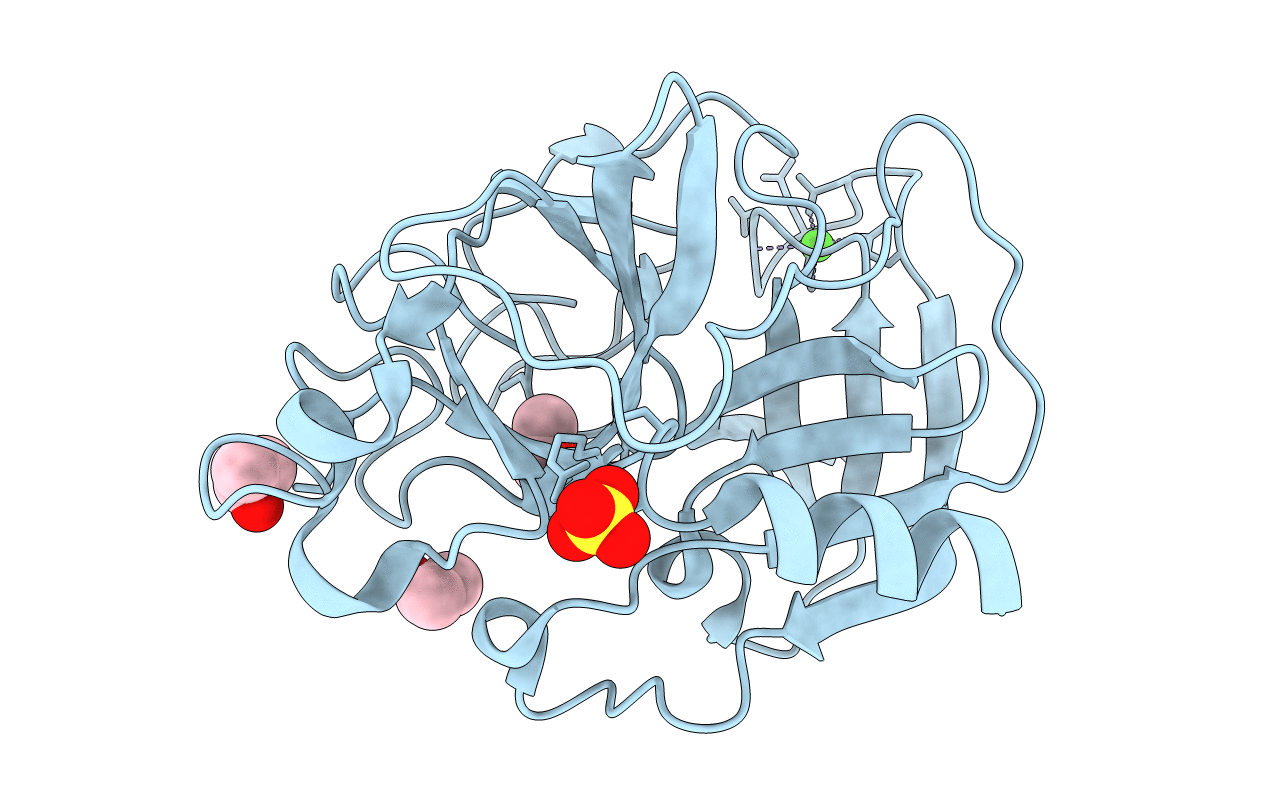 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, IPA |
 |
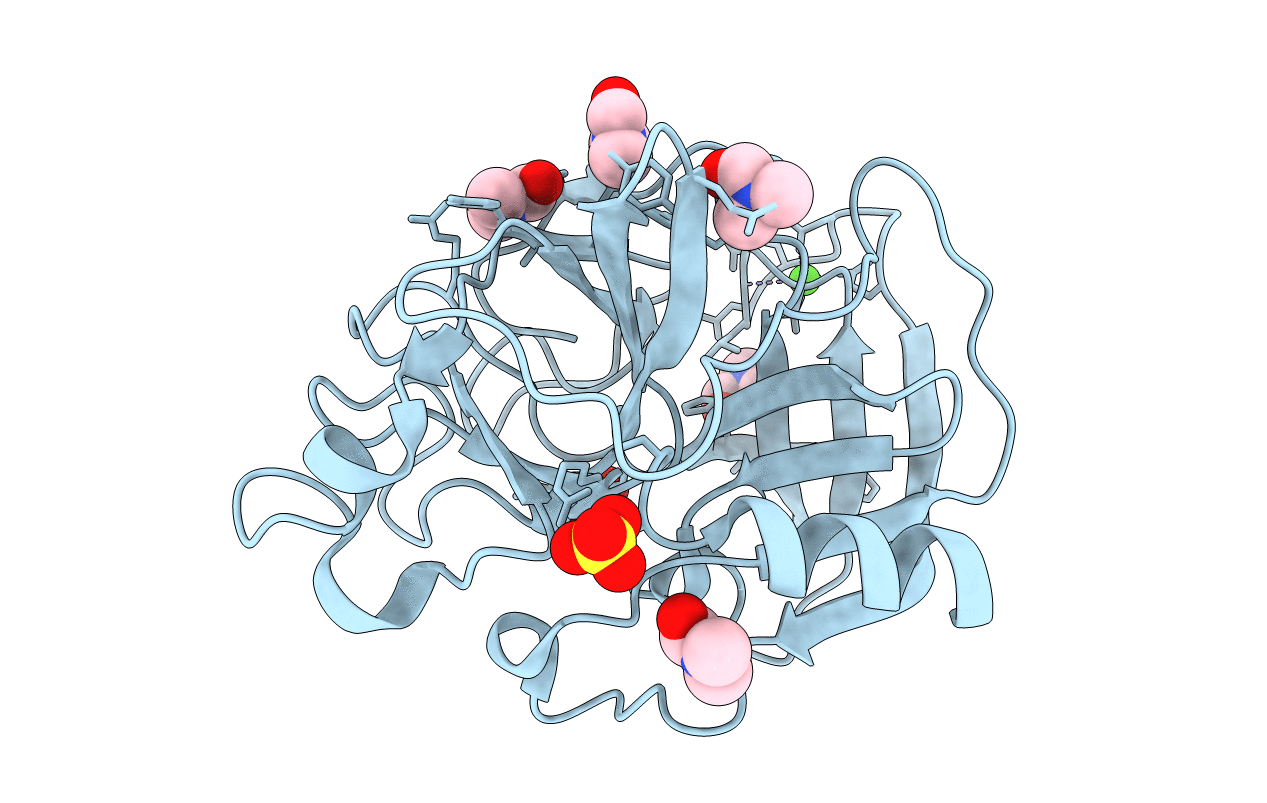
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, IPA |
 |
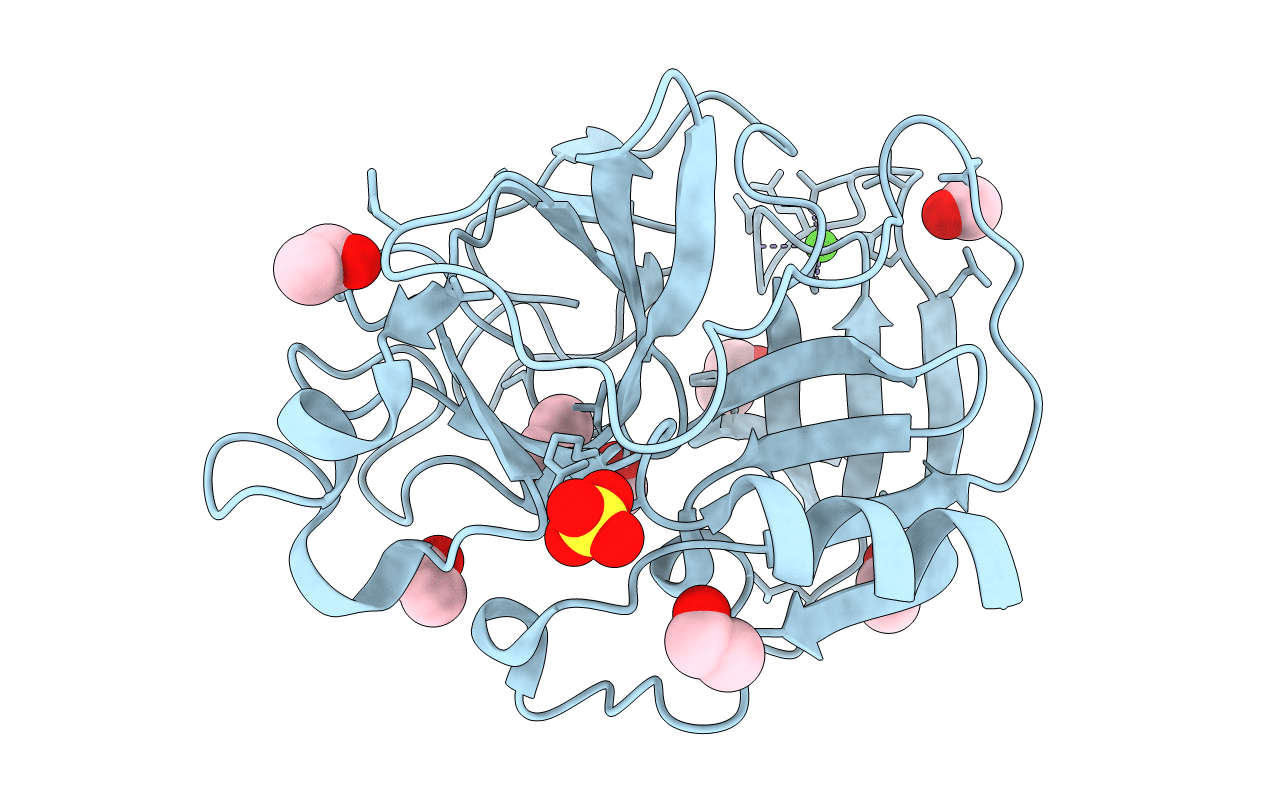
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, DMF |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, EOH |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, 217 |
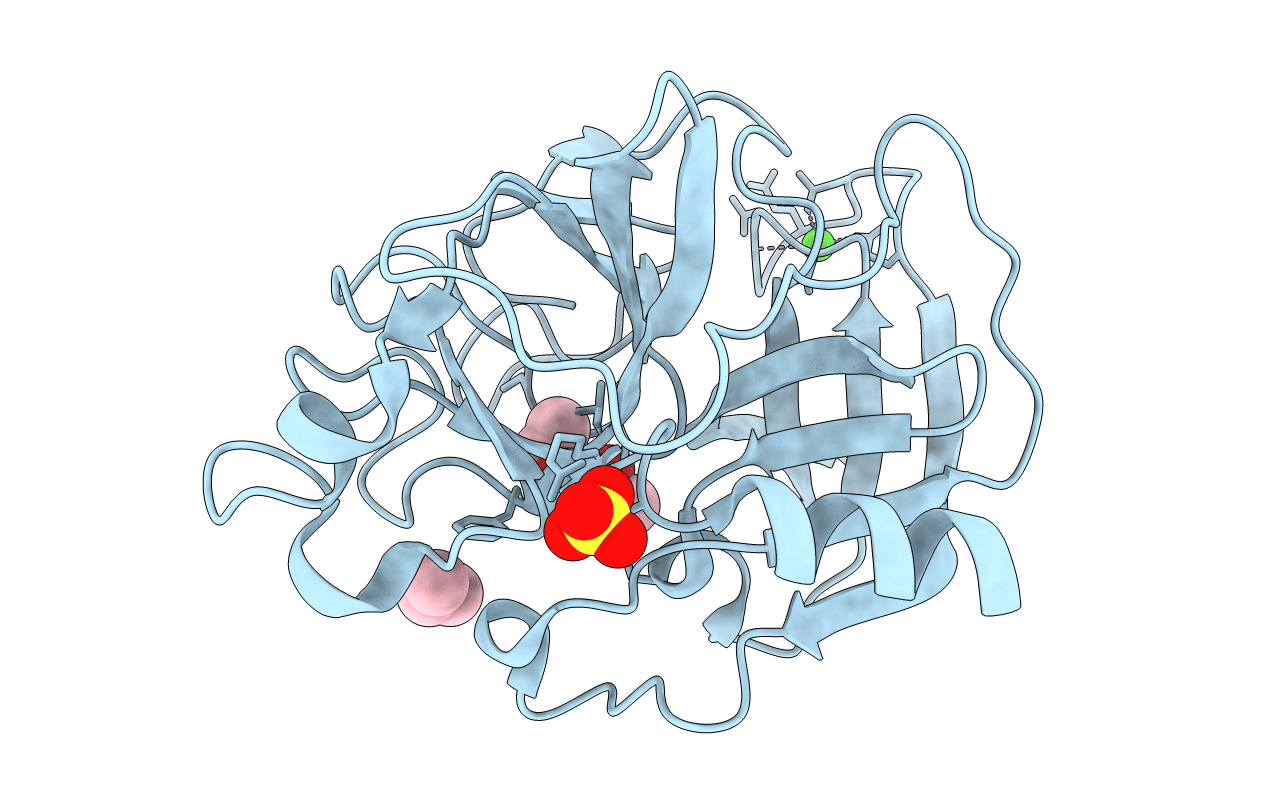 |
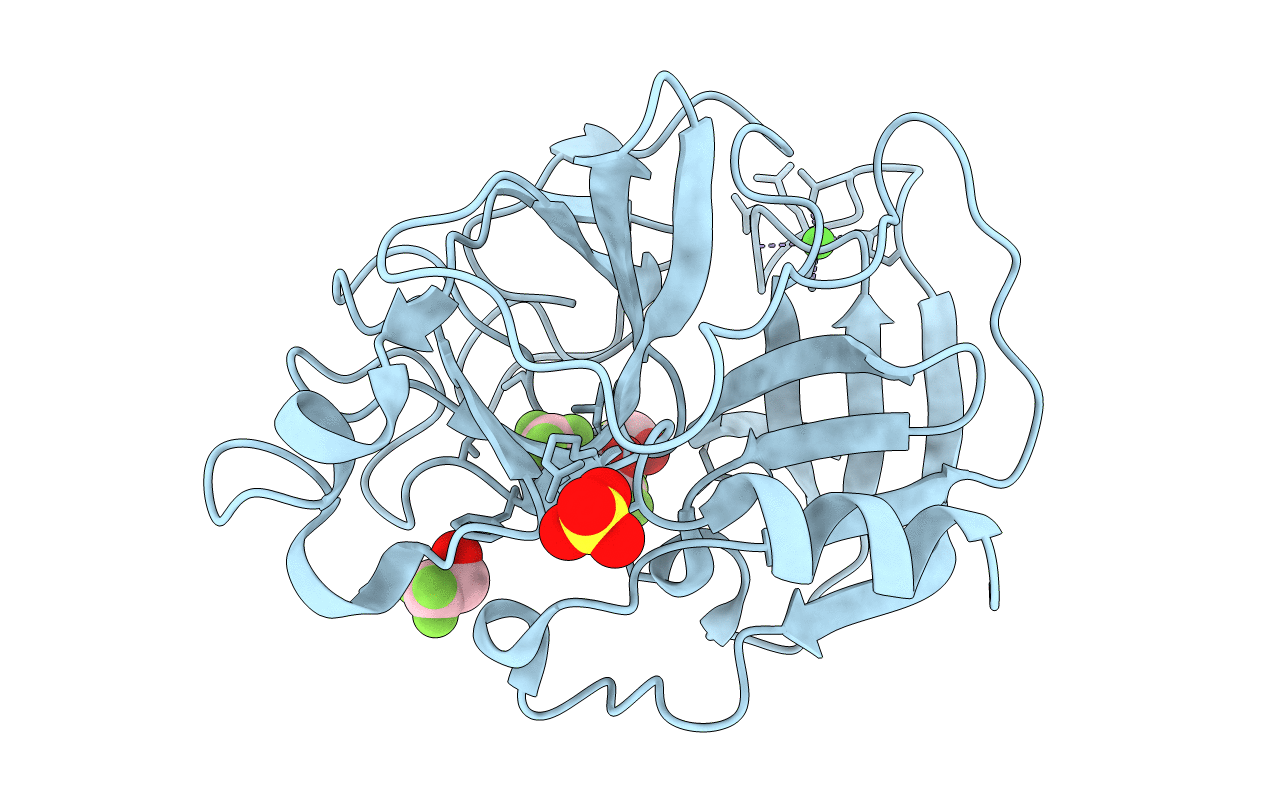
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, IPA |
 |
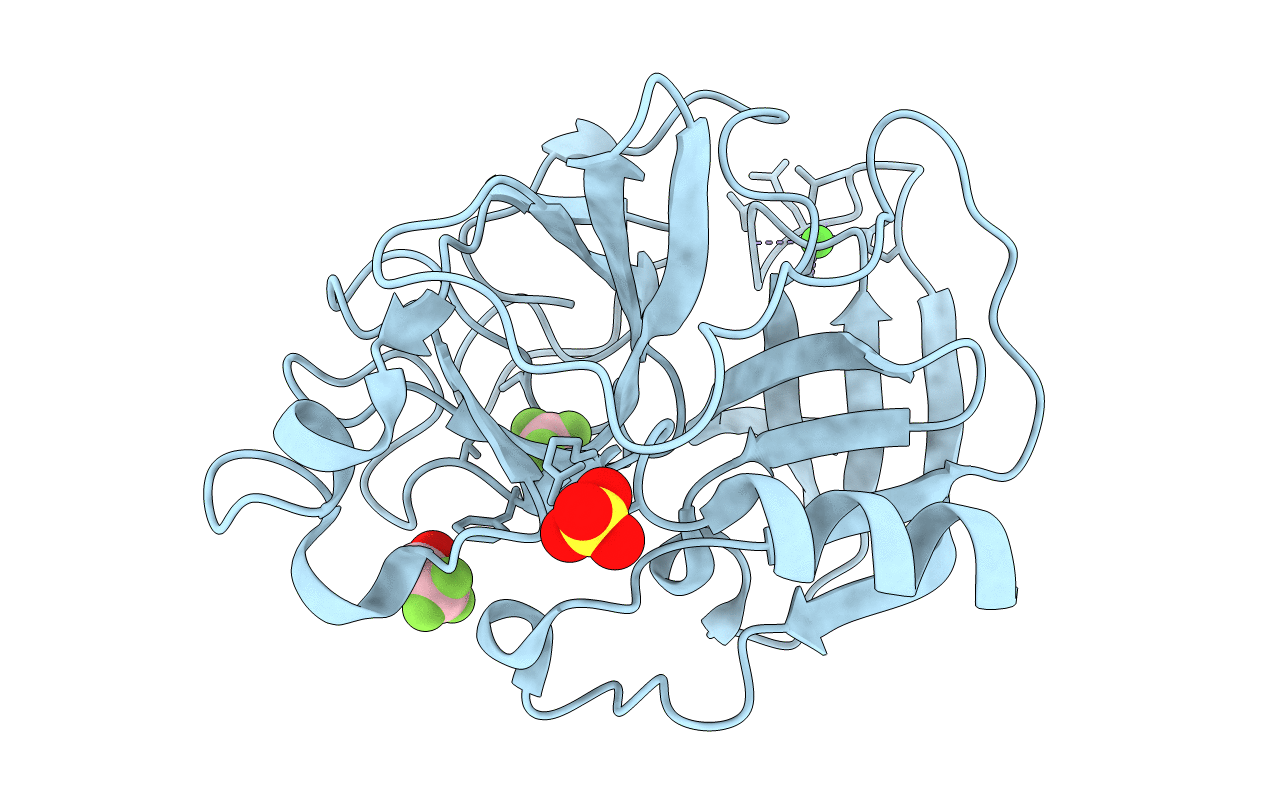
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, ETF |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: CA, SO4, ETF |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-10-26 Classification: HYDROLASE Ligands: ACT |
 |
X-Ray Crystal Structure Of The Hypothetical Phosphotyrosine Phosphatase Mdp-1 Of The Haloacid Dehalogenase Superfamily
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-10-19 Classification: HYDROLASE Ligands: MG, WO4 |
 |
Method: X-RAY DIFFRACTION
Resolution:1.20 Å Release Date: 2004-08-17 Classification: DE NOVO PROTEIN |

