Search Count: 36
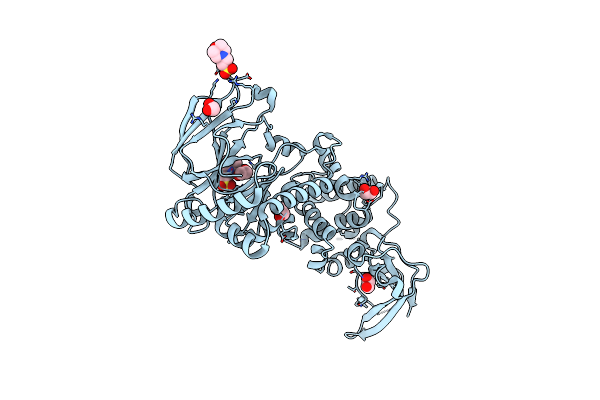 |
The Structure Of The Catalytic Module Of The Metalloprotease Zmpa From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: MES, EDO |
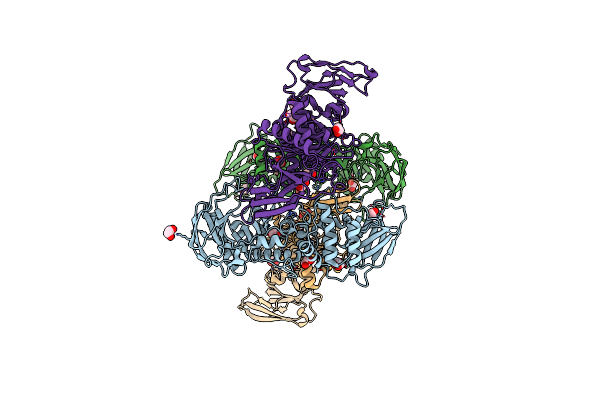 |
The Structure Of The M60 Catalytic Domain From Clostridium Perfringens Zmpc
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN, EDO |
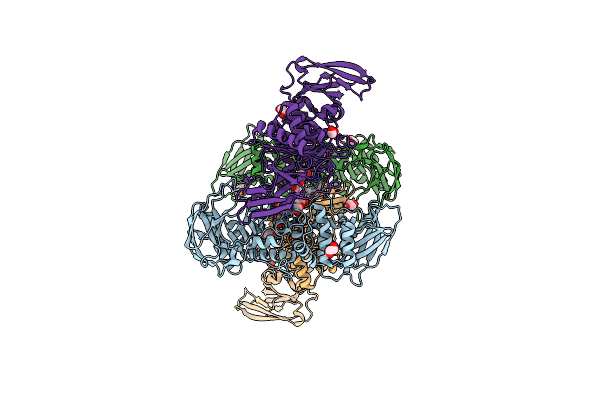 |
The Structure Of The M60 Catalytic Domain From Clostridium Perfringens Zmpc In Complex The Sialyl T Antigen
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN, EDO, SER |
 |
The Structure Of The Cbm32-1, Cbm32-2, And M60 Catalytic Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN |
 |
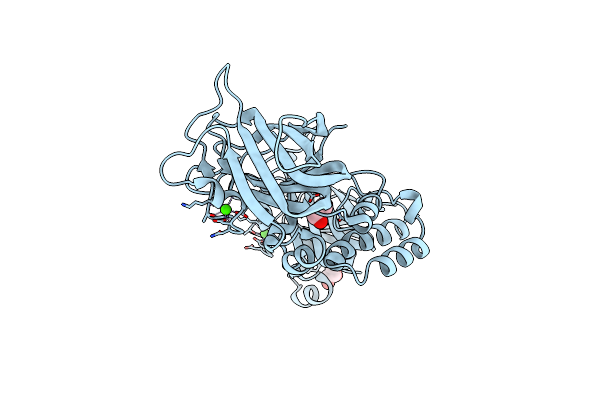
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, A2G, GOL |
 |
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: A2G, CA, GOL |
 |
The Structure Of Cbm51-2 In Complex With Glcnac And Int Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, NAG, EDO, PO4, TRS |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
The Structure Of The M60 Catalytic Domain With The Cbm51-1 And Cbm51-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE |
 |
Structure Of Impa From Pseudomonas Aeruginosa In Complex With An O-Glycopeptide
Organism: Pseudomonas aeruginosa, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-12-16 Classification: PROTEIN BINDING Ligands: ZN, EDO |
 |
Crystal Structure Of Ruminococcus Flavefaciens' Type Iii Complex Containing The Fifth Cohesin From Scaffoldin B And The Dockerin From Scaffoldin A
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-02-28 Classification: PROTEIN BINDING Ligands: CCN, CA |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, EDO |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, EDO, PO4 |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, NI, NA, SER, A2G |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GOL, NA, ZN |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: EDO, ZN |
 |
Zmpb Metallopeptidase In Complex With An O-Glycopeptide (A2,6-Sialylated Core-3 Pentapeptide).
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a), Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, TLA, EPE, EDO |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: EDO, ZN, SER |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / pao1 / 1c / prs 101 / lmg 12228)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: SO4, ZN, EDO |

