Search Count: 15
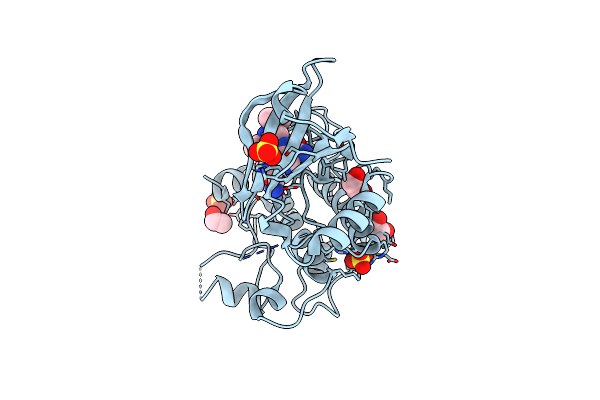 |
Crystal Structure Of Human Calmodulin-Dependent Protein Kinase 1D (Camk1D) Bound To Compound 19 (Cs640)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-11-13 Classification: TRANSFERASE Ligands: SO4, EDO, M92 |
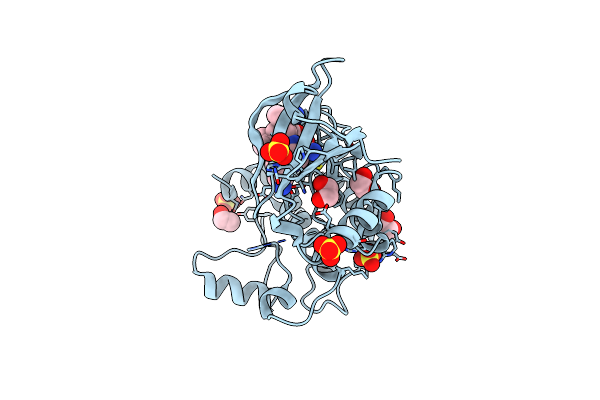 |
Crystal Structure Of Human Calmodulin-Dependent Protein Kinase 1D (Camk1D) Bound To Compound 18 (Cs587)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-11-13 Classification: TRANSFERASE Ligands: SO4, EDO, M8Z |
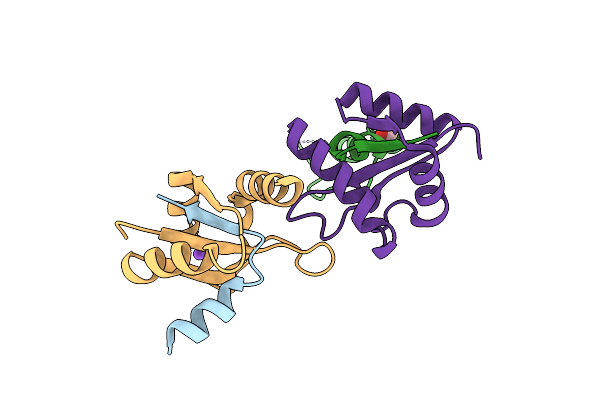 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
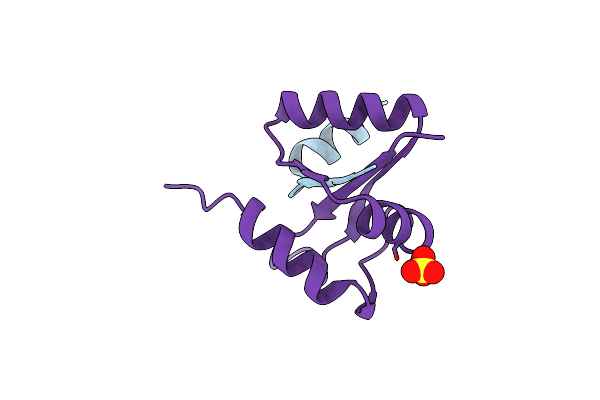 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Crystal Structure Of Escu C-Terminal Domain With N262D Mutation, Space Group P 41 21 2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Crystal Structure Of Escu C-Terminal Domain With N262D Mutation, Space Group P 21 21 21
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT Ligands: SO4, CYS, MPD |
 |
Crystal Structure Of The Escu C-Terminal Domain With P263A Mutation,Space Group P 1 21 1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-22 Classification: MEMBRANE PROTEIN, PROTEIN TRANSPORT Ligands: PRO |

