Search Count: 25
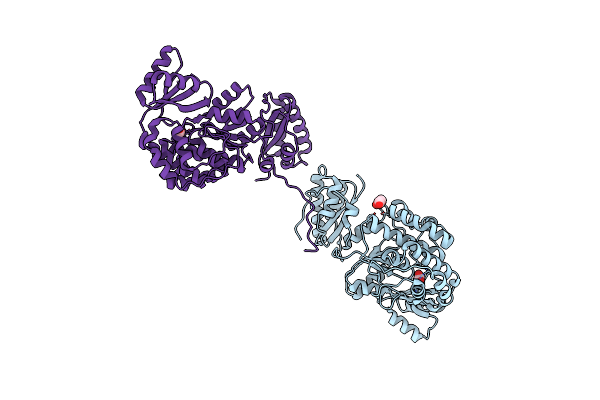 |
Structure Of The S726F Mutant Of Acyltransferase Domain Of Mycocerosic Acid Synthase From Mycobacterium Tuberculosis
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-16 Classification: TRANSFERASE Ligands: ACT, GOL |
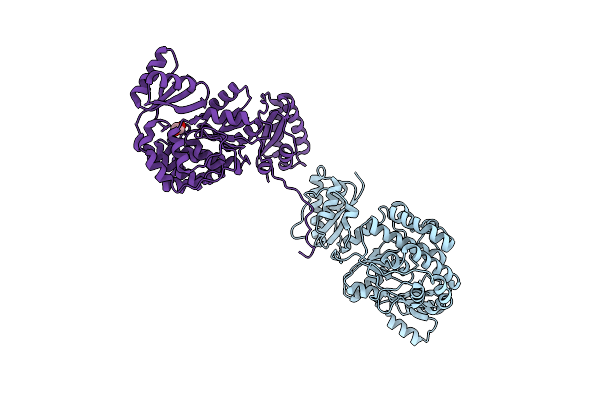 |
Structure Of The M624V-S726F Mutant Of Acyltransferase Domain Of Mycocerosic Acid Synthase From Mycobacterium Tuberculosis Soaked With Methylmalonyl Coenzyme A
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-16 Classification: TRANSFERASE Ligands: EDO |
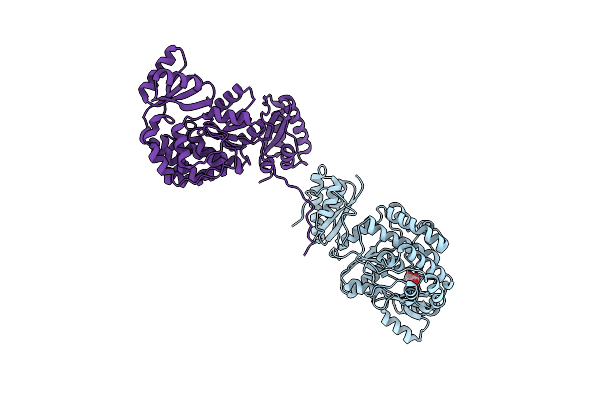 |
Structure Of The Acyltransferase Domain Of Mycocerosic Acid Synthase From Mycobacterium Tuberculosis Acylated Methylmalonyl-Coenzymea
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-12-16 Classification: TRANSFERASE Ligands: DXX |
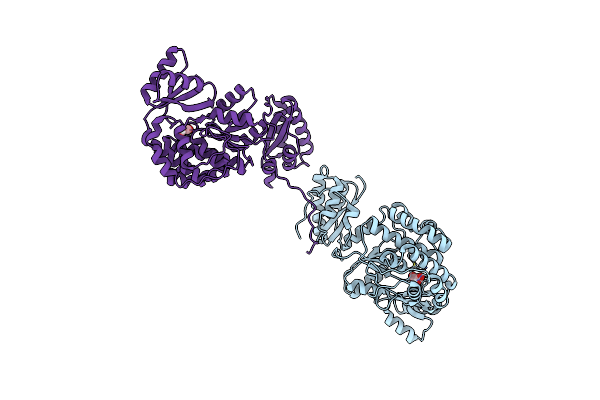 |
Structure Of The S726F Mutant Of Acyltransferase Domain Of Mycocerosic Acid Synthase From Mycobacterium Tuberculosis Acylated With Malonyl-Coenzyme A
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-12-16 Classification: TRANSFERASE Ligands: MLA |
 |
Structure Of The S726F Mutant Of Acyltransferase Domain Of Mycocerosic Acid Synthase From Mycobacterium Tuberculosis Acylated With Methylmalonyl-Coenzyme A
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-12-16 Classification: TRANSFERASE Ligands: DXX, EDO |
 |
Structure Of The Acyltransferase Domain Of Mycocerosic Acid Synthase From Mycobacterium Tuberculosis
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-12-09 Classification: TRANSFERASE Ligands: EDO |
 |
Acyltransferase Domain Of The Polyketide Synthase Ppsc Of Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-09 Classification: TRANSFERASE Ligands: GOL, SO4, SCN, NA |
 |
Structure Of The Of Acyltransferase Domain Of Phenolphthiocerol/Phtiocerol Synthase A From Mycobacterium Bovis (Bcg)
Organism: Mycobacterium bovis (strain bcg / pasteur 1173p2)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-09 Classification: TRANSFERASE |
 |
Organism: Bacillus brevis
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2014-03-19 Classification: ANTIBIOTIC Ligands: MPD, MOH |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Umabf62A From Ustilago Maidys
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, GOL, TRS, 1PE |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Umabf62A From Ustilago Maydis In Complex With L-Arabinofuranose
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, FUB, AHR, TRS |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina In Complex With Cellotriose
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, TRS, 1PE, EPE |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, EPE, TRS, 1PE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-06 Classification: Signaling Protein/Antibiotic Ligands: TLA, CL, N1L, MAN, M12 |
 |
New Strategy To Analyze Structures Of Glycopeptide Antibiotic-Target Complexes
Organism: Escherichia coli k-12, Amycolatopsis lurida
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-06-06 Classification: SUGAR BINDING PROTEIN/ANTIBIOTIC Ligands: IPA, SO4, RST, MAN |
 |
New Strategy To Analyze Structures Of Glycopeptide Antibiotic-Target Complexes
Organism: Enterobacteria phage t4, Streptomyces orientalis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-06-06 Classification: HYDROLASE/ANTIBIOTIC Ligands: MPD, MRD, PO4, TRS, CL, IPA |
 |
Crystal Structure Of The Ligand Binding Domain Of The Retinoid X Receptor Alpha In Complex With 3-(2'-Methoxy)-Tetrahydronaphtyl Cinnamic Acid And A Fragment Of The Coactivator Tif-2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-10-09 Classification: HORMONE RECEPTOR Ligands: 3TN |
 |
Crystal Structure Of The Ligand Binding Domain Of The Retinoid X Receptor Alpha In Complex With 3-(2'-Ethoxy)-Tetrahydronaphtyl Cinnamic Acid And A Fragment Of The Coactivator Tif-2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-10-09 Classification: HORMONE RECEPTOR Ligands: 4TN |
 |
Crystal Structure Of The Ligand Binding Domain Of The Retinoid X Receptor Alpha In Complex With 3-(2'-Propoxy)-Tetrahydronaphtyl Cinnamic Acid And A Fragment Of The Coactivator Tif-2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-10-09 Classification: HORMONE RECEPTOR Ligands: 5TN |
 |
Crystal Structure Of The Pig Pancreatic Alpha-Amylase Complexed With Malto-Oligosaacharides Under The Effect Of The Chloride Ion
Organism: sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2005-03-15 Classification: HYDROLASE Ligands: CL, CA, EDO |

