Search Count: 18
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Dionaea muscipula
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: PLM |
 |
Organism: Dionaea muscipula
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: PLM |
 |
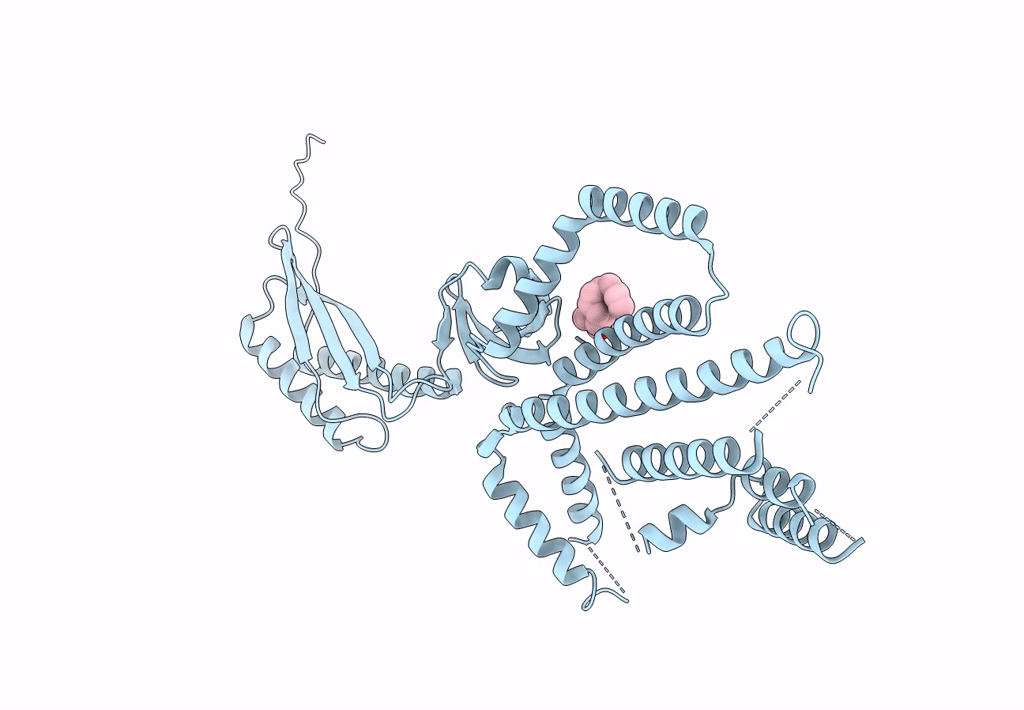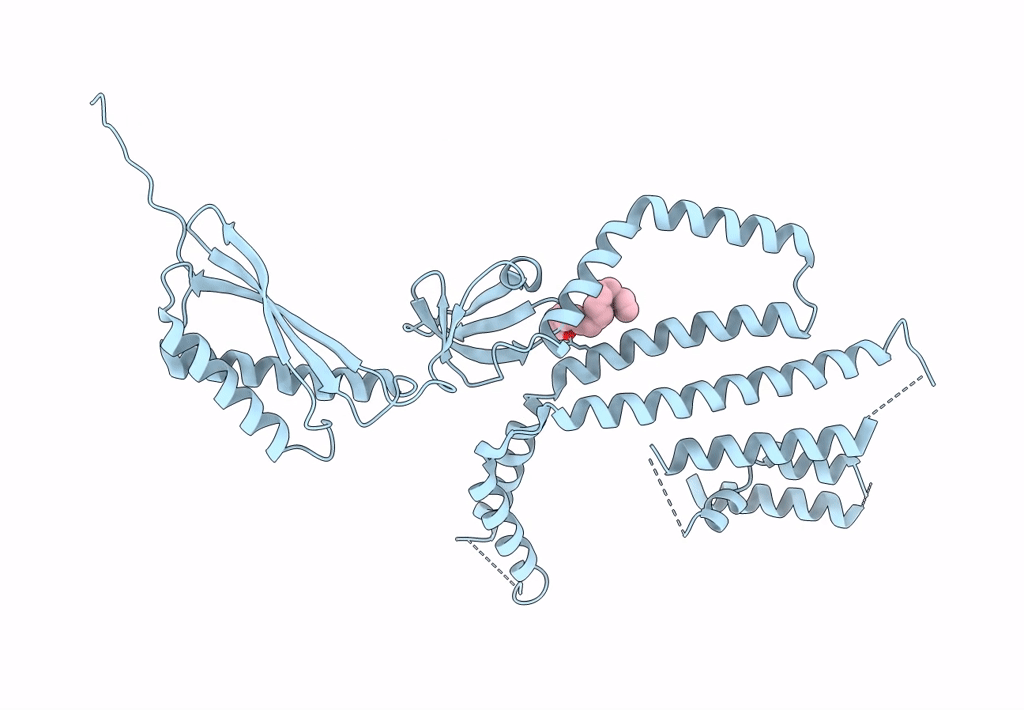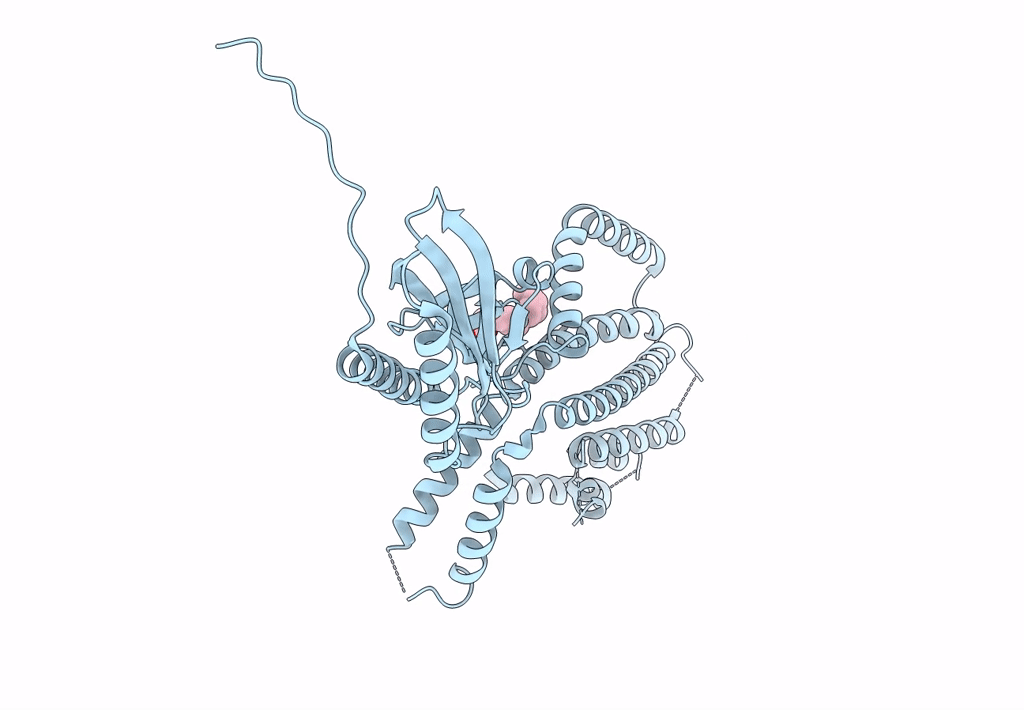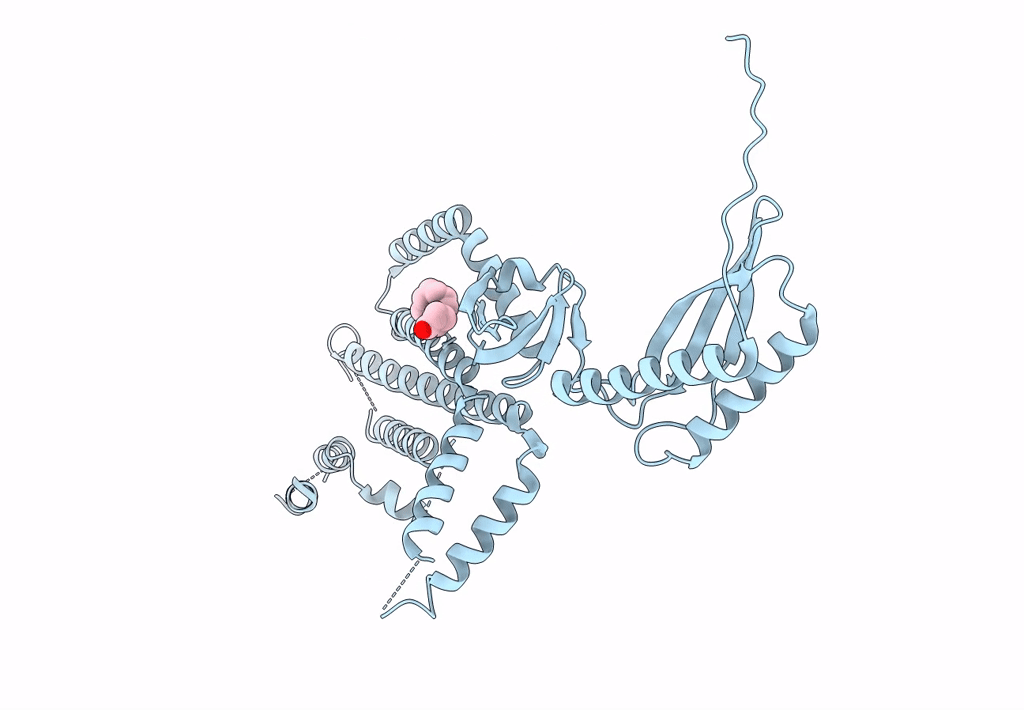
Structure Of Mechanosensitive Ion Channel Flycatcher1 Protomer In 'Down' Conformation In Gdn
Organism: Dionaea muscipula
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: PLM |
 |
Structure Of Mechanosensitive Ion Channel Flycatcher1 Protomer In 'Up' Conformation In Gdn
Organism: Dionaea muscipula
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: PLM |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2018-11-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2018-11-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN |
 |
Quisqualate Bound To The D655A Mutant Of The Ligand Binding Domain Of Glua3
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-30 Classification: TRANSPORT PROTEIN/AGONIST Ligands: QUS, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2012-05-30 Classification: TRANSPORT PROTEIN/AGONIST Ligands: KAI, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2012-05-30 Classification: TRANSPORT PROTEIN/AGONIST Ligands: KAI, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2012-05-30 Classification: TRANSPORT PROTEIN/SUBSTRATE Ligands: GLU, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2012-05-30 Classification: TRANSPORT PROTEIN/AGONIST Ligands: KAI, ZN |
 |
Quisqualate Bound To The D655A Mutant Of The Ligand Binding Domain Of Glua3
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2012-05-23 Classification: TRANSPORT PROTEIN/AGONIST Ligands: QUS, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-05-16 Classification: TRANSPORT PROTEIN/AGONIST Ligands: CNI, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2012-05-16 Classification: TRANSPORT PROTEIN/AGONIST Ligands: KAI, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-05-16 Classification: TRANSPORT PROTEIN/AGONIST Ligands: QUS, ZN |

