Search Count: 14
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Ncl-00023818
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2021-07-14 Classification: CHAPERONE Ligands: PG4, AN2, GOL, PYZ, PO4, CL, IOD |
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Z321318226
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2021-06-02 Classification: CHAPERONE Ligands: GOL, PG4, AN2, JHJ, PO4, CL |
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Ncl-00023823
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2021-06-02 Classification: CHAPERONE Ligands: GOL, PG4, AN2, HEW, PO4, CL, MG |
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Z1203107138
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2021-03-17 Classification: CHAPERONE Ligands: AN2, GOL, PG4, PO4, JH1 |
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Z287256168
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2021-03-10 Classification: CHAPERONE Ligands: GOL, PG6, AN2, PO4, CL, SYQ |
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Z396380540
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2021-03-10 Classification: CHAPERONE Ligands: GOL, AN2, PG4, PO4, CL, HR5 |
 |
Plasmodium Falciparum Hsp70-X Chaperone Nucleotide Binding Domain In Complex With Z1827898537
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2021-03-10 Classification: CHAPERONE Ligands: GOL, PG4, AN2, PO4, CL, WM7 |
 |
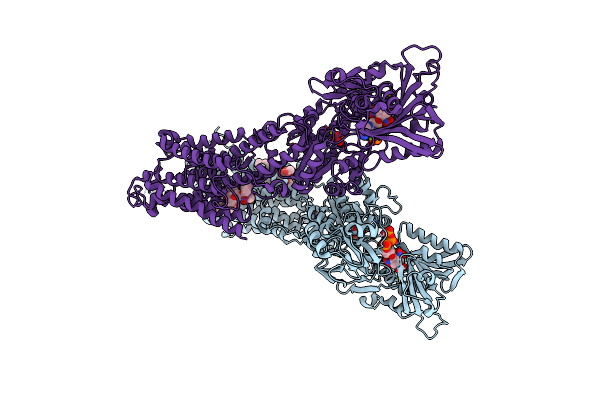
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: UVC |
 |
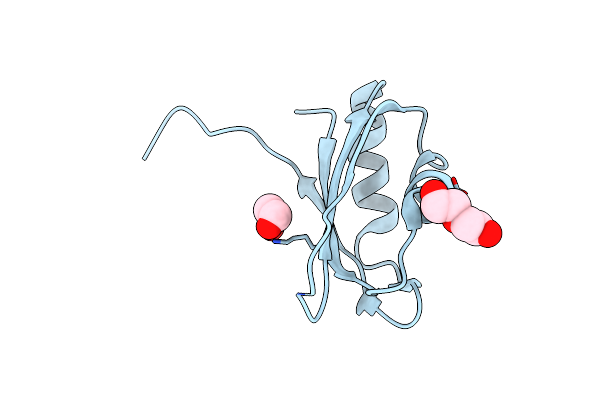
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: 128, TG1, VN4, MG, CL, K |
 |
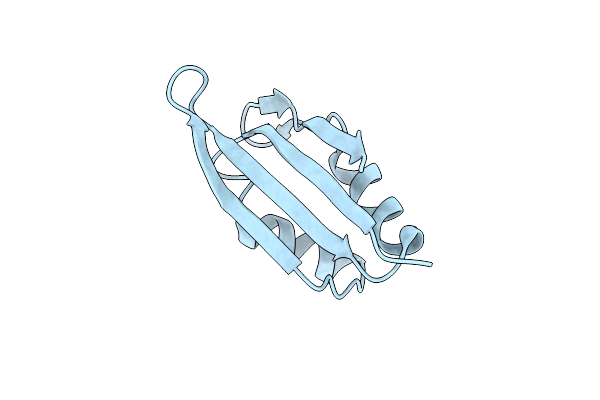
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-12-12 Classification: RNA BINDING PROTEIN Ligands: PEG |
 |
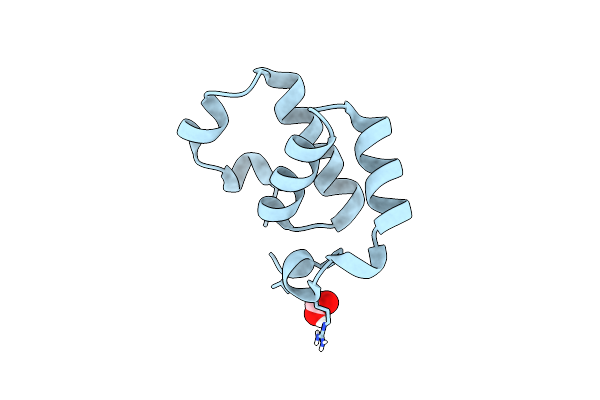
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-11-21 Classification: RNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: FMT |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-08-17 Classification: RNA BINDING PROTEIN Ligands: CIT, NO3, NA |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2016-08-17 Classification: RNA BINDING PROTEIN Ligands: BME, SO4 |

