Search Count: 25
 |
Organism: Escherichia coli (strain k12), Rna interference vector pbsk-gus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-02-05 Classification: HYDROLASE Ligands: MG |
 |
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-02-05 Classification: HYDROLASE |
 |
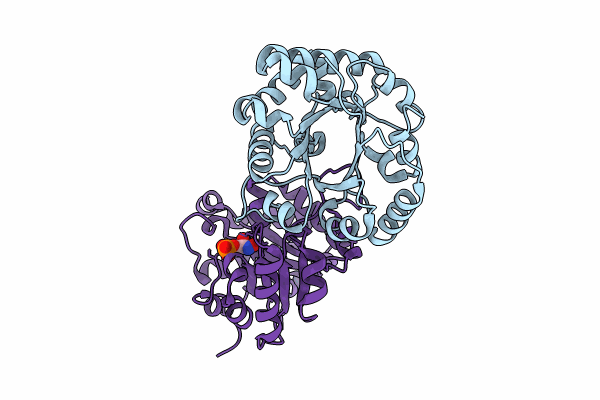
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
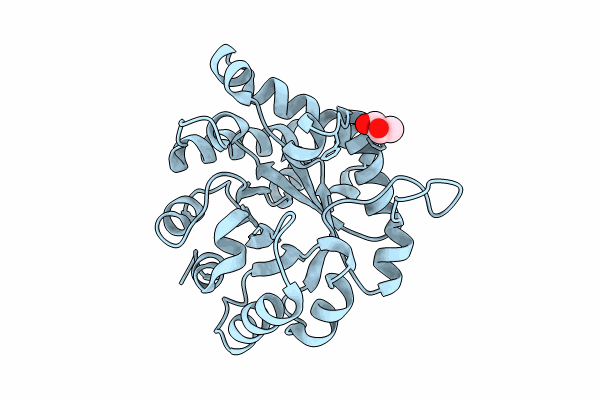
Crystal Structure Of The Worst Case Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH |
 |
Crystal Structure Of The Ancestral Triosephosphate Isomerase Reconstruction Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: GOL |
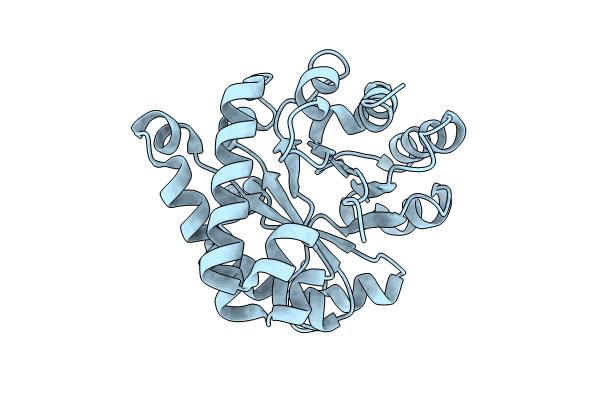 |
Crystal Structure Of The Reconstruction Of The Worst Case Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH, ACY, PGE, ACT, NA, EDO, PEG, CL |
 |
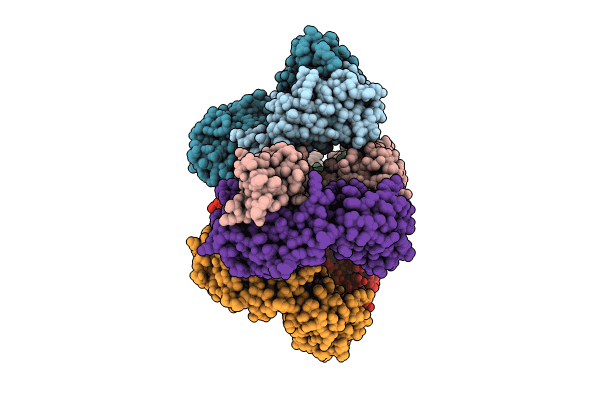
Crystal Structure Of The Worst Case Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: F |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: SUGAR BINDING PROTEIN |
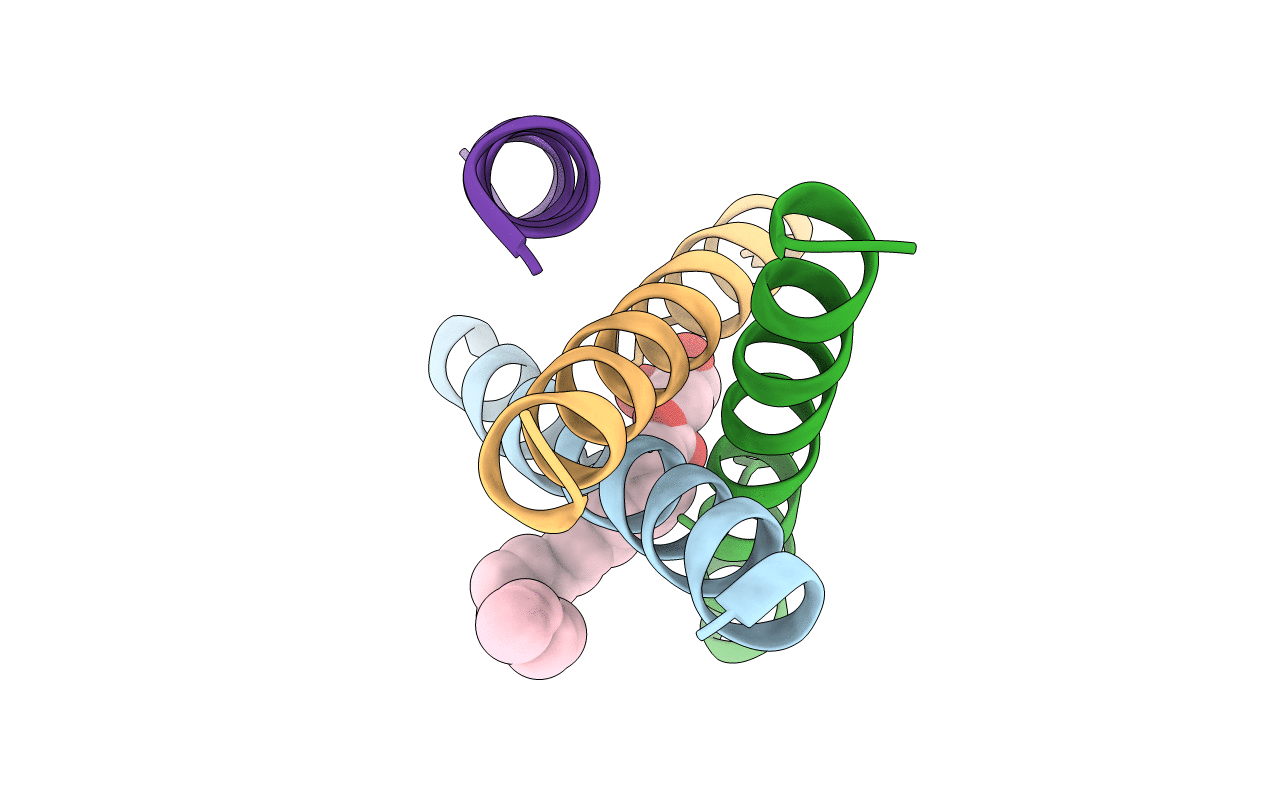 |
De Novo Designed Receptor Transmembrane Domains Enhance Car-T Cytotoxicity And Attenuate Cytokine Release
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-03-31 Classification: BIOSYNTHETIC PROTEIN Ligands: OLB |
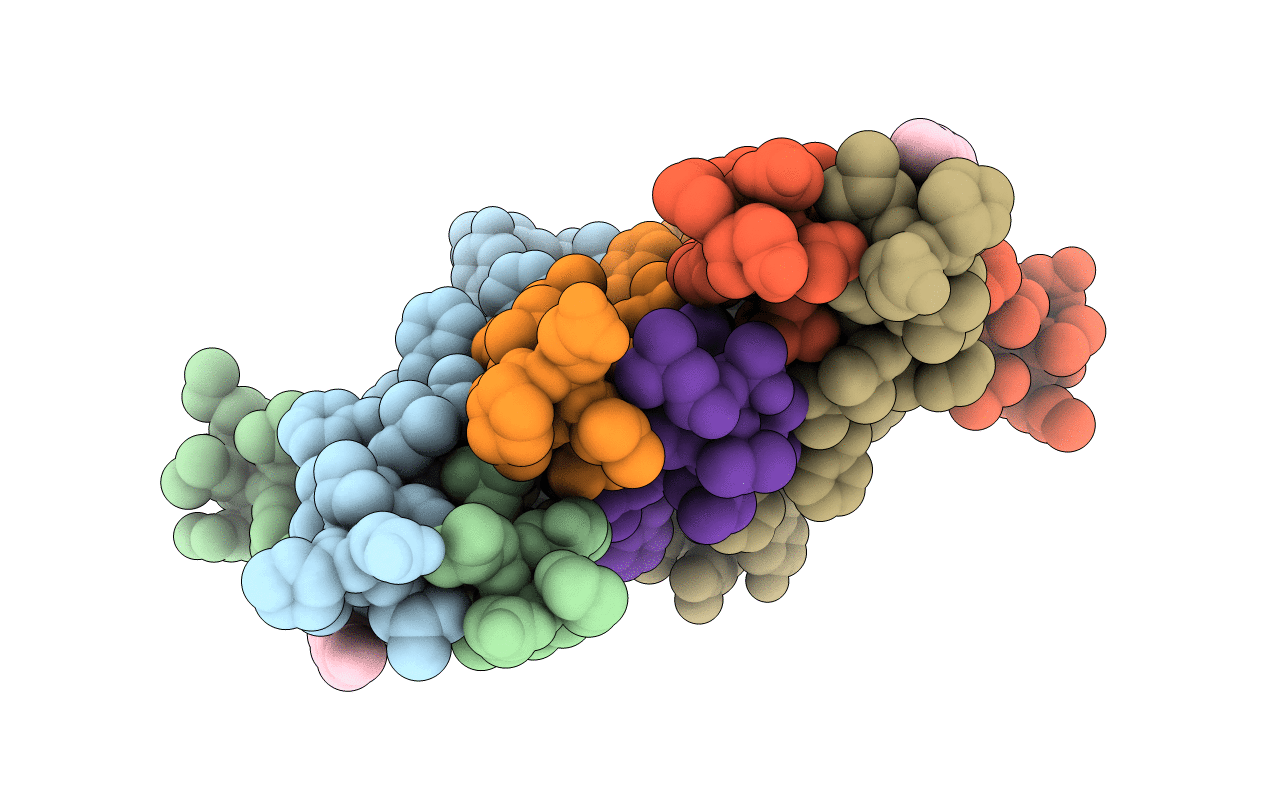 |
De Novo Designed Receptor Transmembrane Domains Enhance Car-T Cytotoxicity And Attenuate Cytokine Release
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-03-31 Classification: BIOSYNTHETIC PROTEIN Ligands: OLB |
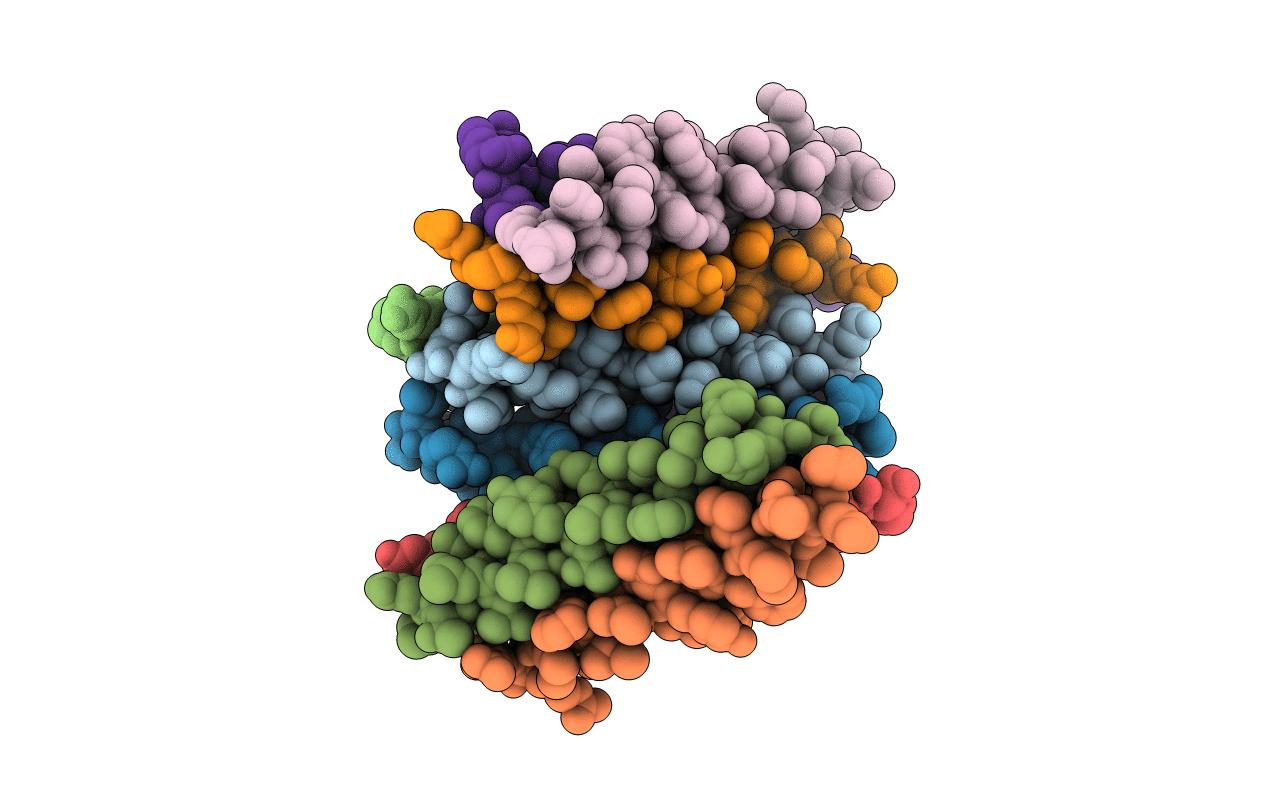 |
De Novo Designed Receptor Transmembrane Domains Enhance Car-T Cytotoxicity And Attenuate Cytokine Release
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2021-03-31 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of The Human Respiratory Syncytial Virus M2-1 Protein In Complex With A Short Positive-Sense Gene-End Rna
Organism: Human respiratory syncytial virus a (strain a2), Human respiratory syncytial virus a2
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-08-05 Classification: METAL BINDING PROTEIN/RNA Ligands: ZN |
 |
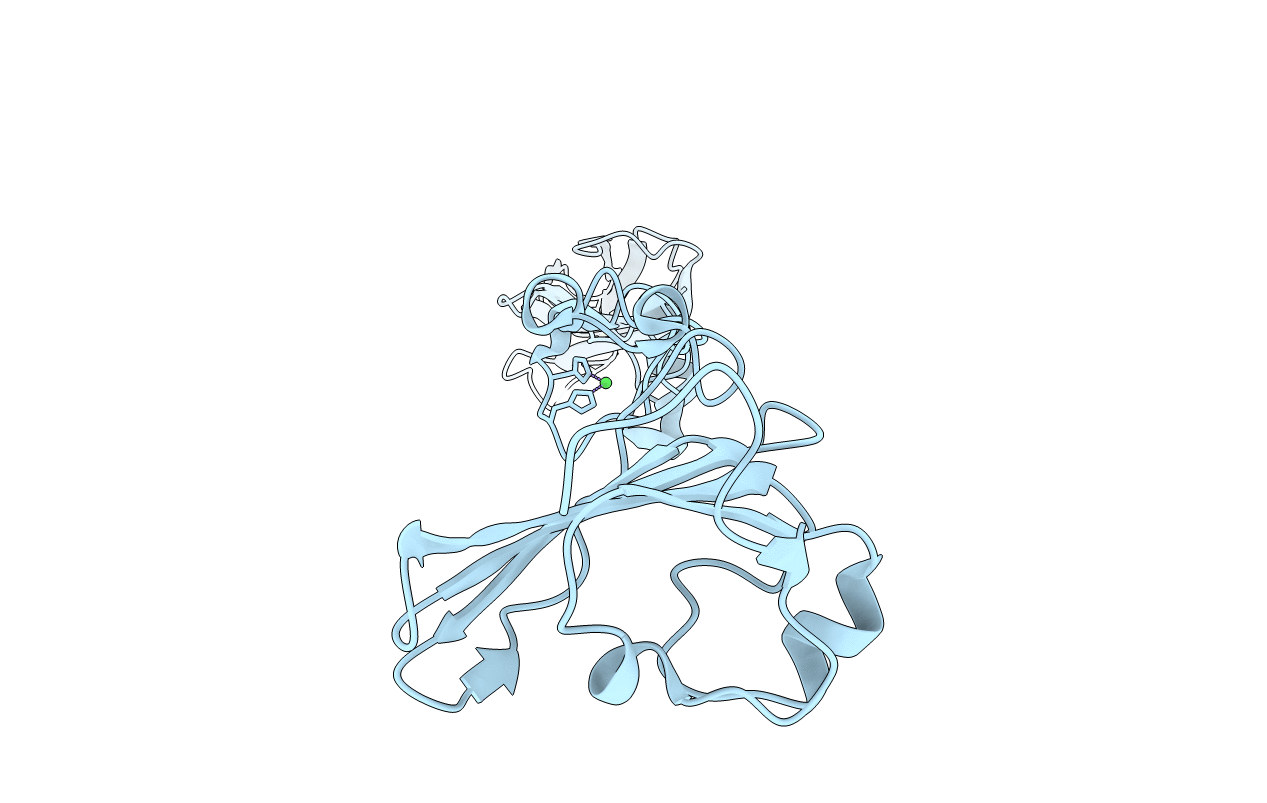
Selenomethionine Structure Of N-Truncated R2-Type Pyocin Tail Fiber At 2.6 Angstrom Resolution
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2019-02-20 Classification: VIRAL PROTEIN Ligands: NI |
 |
Structure Of N-Truncated R2-Type Pyocin Tail Fiber At 2.6 Angstrom Resolution
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2019-02-20 Classification: VIRAL PROTEIN Ligands: MG, NI |
 |
Structure Of N-Truncated R1-Type Pyocin Tail Fiber At 1.7 Angstrom Resolution
Organism: Pseudomonas aeruginosa lesb58
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-20 Classification: VIRAL PROTEIN Ligands: MG, GOL |
 |
Cryo-Em Structure Of Human Parainfluenza Virus Type 3 (Hpiv3) In Complex With Antibody Pia174
Organism: Human parainfluenza virus 3, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.30 Å Release Date: 2018-11-14 Classification: VIRAL PROTEIN/immune system |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2018-06-06 Classification: RNA BINDING PROTEIN |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-04-04 Classification: ISOMERASE |

