Search Count: 20
 |
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-05-17 Classification: HYDROLASE/INHIBITOR Ligands: ZN, CAC |
 |
Crystal Structure Of Fi-Cmcase From Aspergillus Aculeatus F-50 In Complex With Cellotetrose
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-05-17 Classification: HYDROLASE/INHIBITOR Ligands: SO4 |
 |
Crystal Structure Of Fi-Cmcase From Aspergillus Aculeatus F-50 In Complex With Cellobiose
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2017-05-17 Classification: HYDROLASE/INHIBITOR Ligands: SO4, EPE |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-12-21 Classification: HYDROLASE Ligands: CA, PGE, PEG, NAG |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2016-12-21 Classification: HYDROLASE Ligands: CA, NAG, PGE |
 |
Crystal Structure Of A Beta-1,4-Endoglucanase Mutant From Aspergillus Niger In Complex With Sugar
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-12-21 Classification: HYDROLASE Ligands: PGE, NAG, CA |
 |
The Truncated Fibrobacter Succinogenes 1,3-1,4-Beta-D-Glucanase V18Y/W203Y In Apo-Form
Organism: Fibrobacter succinogenes
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2012-02-15 Classification: HYDROLASE Ligands: TRS, CA |
 |
The Truncated Fibrobacter Succinogenes 1,3-1,4-Beta-D-Glucanase V18Y/W203Y In Complex With Cellotetraose (Cellobiose Density Was Observed)
Organism: Fibrobacter succinogenes
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2012-02-15 Classification: HYDROLASE Ligands: TRS, CA |
 |
Crystal Structures Of Thermotoga Maritima Cel5A In Complex With Cellobiose Substrate, Mutant Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-08-17 Classification: HYDROLASE Ligands: BGC |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-08-10 Classification: HYDROLASE |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-10 Classification: HYDROLASE |
 |
Crystal Structures Of Thermotoga Maritima Cel5A In Complex With Mannotriose Substrate
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2011-08-10 Classification: HYDROLASE |
 |
Diverse Substrates Recognition Mechanism Revealed By Thermotoga Maritima Cel5A Structures In Complex With Cellobiose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2011-08-10 Classification: HYDROLASE Ligands: BGC |
 |
Diverse Substrates Recognition Mechanism Revealed By Thermotoga Maritima Cel5A Structures In Complex With Mannotriose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2011-08-10 Classification: HYDROLASE |
 |
Diverse Substrates Recognition Mechanism Revealed By Thermotoga Maritima Cel5A Structures In Complex With Cellotetraose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-10 Classification: HYDROLASE |
 |
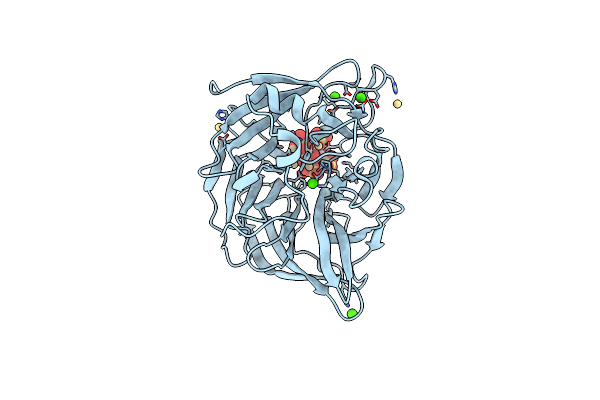
Crystal Structures Of Bacillus Subtilis Alkaline Phytase In Complex With Ca2+, Co2+, Ni2+, Mg2+ And Myo-Inositol Hexasulfate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2011-04-13 Classification: Hydrolase/HYDROLASE INHIBITOR Ligands: IHS, CA |
 |
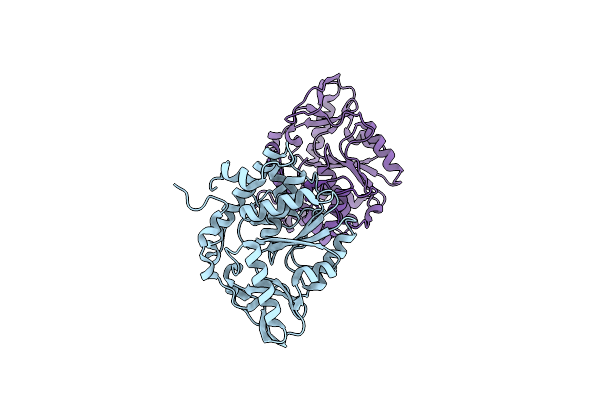
Crystal Structures Of Bacillus Subtilis Alkaline Phytase In Complex With Ca2+, Cd2+, Co2+, Ni2+, Mg2+ And Myo-Inositol Hexasulfate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2011-04-13 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: IHS, CA, CD |
 |
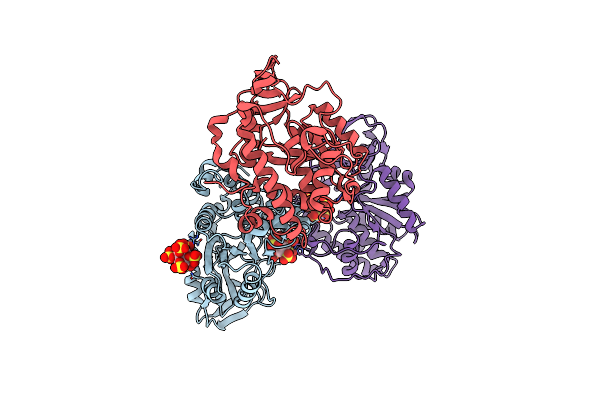
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-11-09 Classification: HYDROLASE |
 |
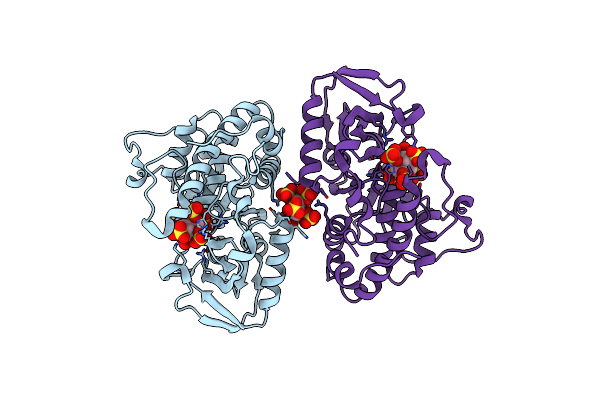
Crystal Structure Of Selenomonas Ruminantium Phytase Complexed With Persulfated Phytate In The C2221 Crystal Form
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-11-09 Classification: HYDROLASE Ligands: IHS |
 |
Crystal Structure Of Selenomonas Ruminantium Phytase Complexed With Persulfated Phytate
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-11-09 Classification: HYDROLASE Ligands: IHS |

