Search Count: 54
 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: Y41F-L68F Double Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: L68Y Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Hemimethylated Dna: L68F Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: L68F Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
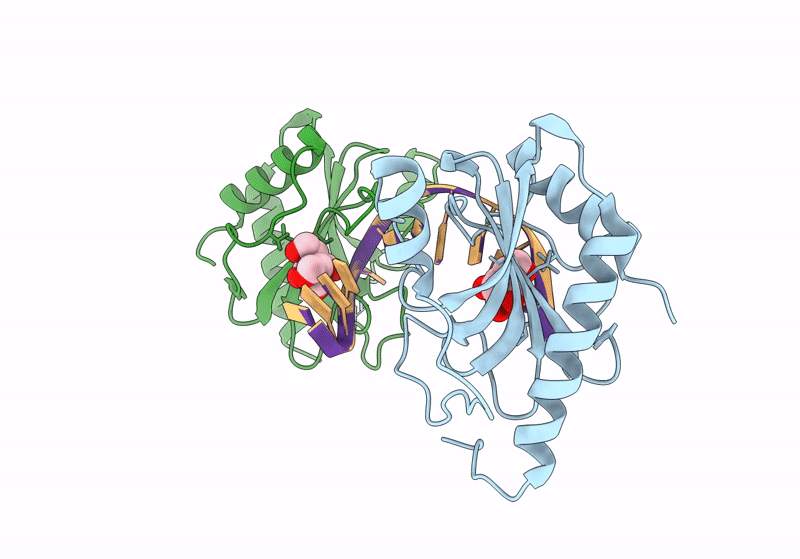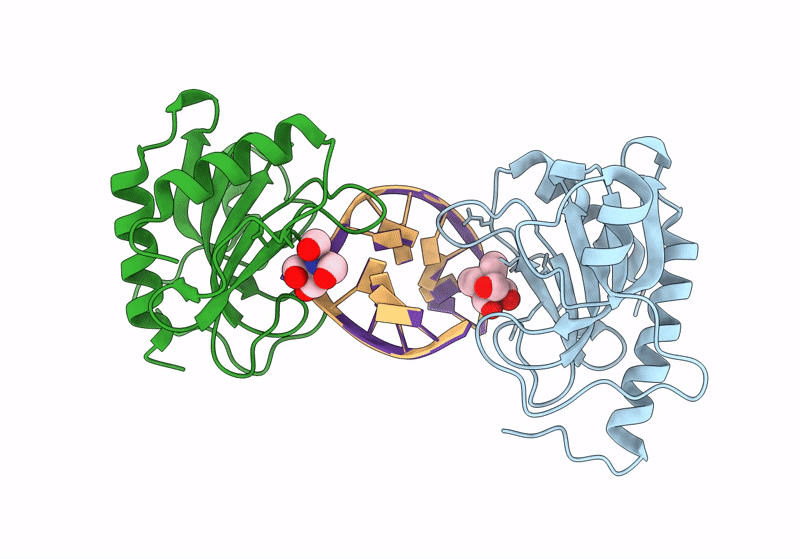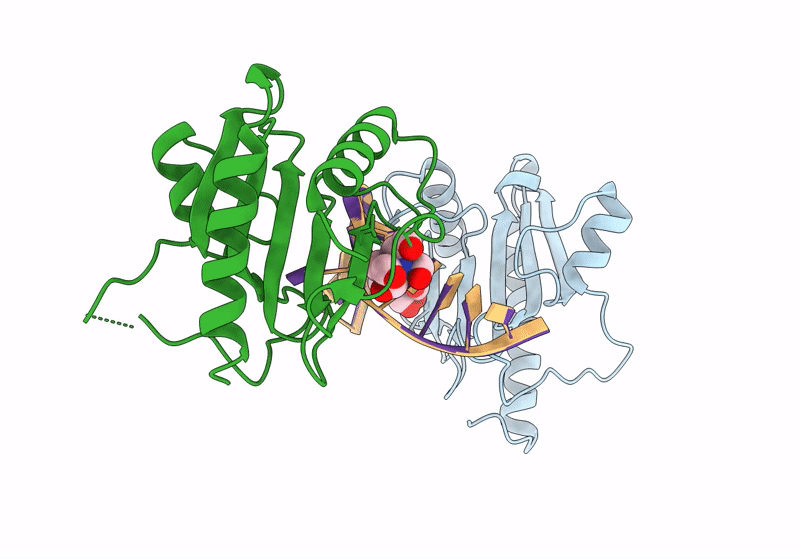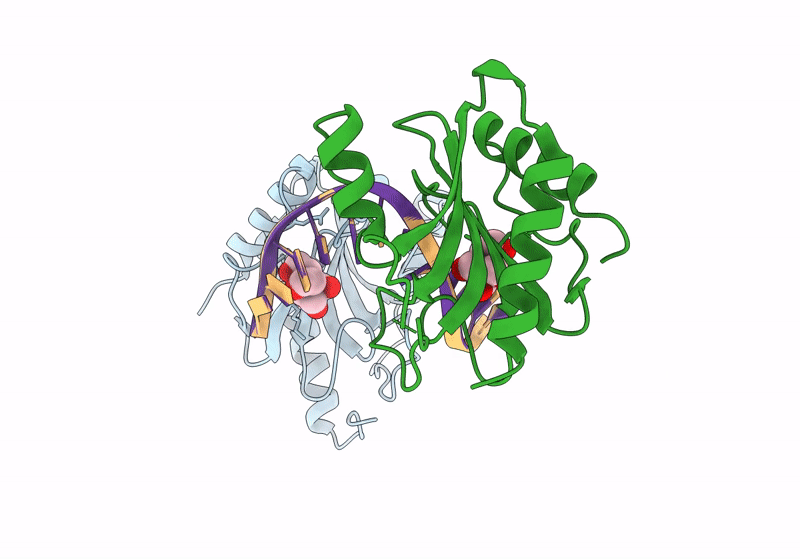 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: Y41F-L68Y Double Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
 |
Crystal Structure Of Wild-Type Human Formylglycine Generating Enzyme Bound To Cu(I)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2023-08-23 Classification: OXIDOREDUCTASE Ligands: CU1, NAG, CA |
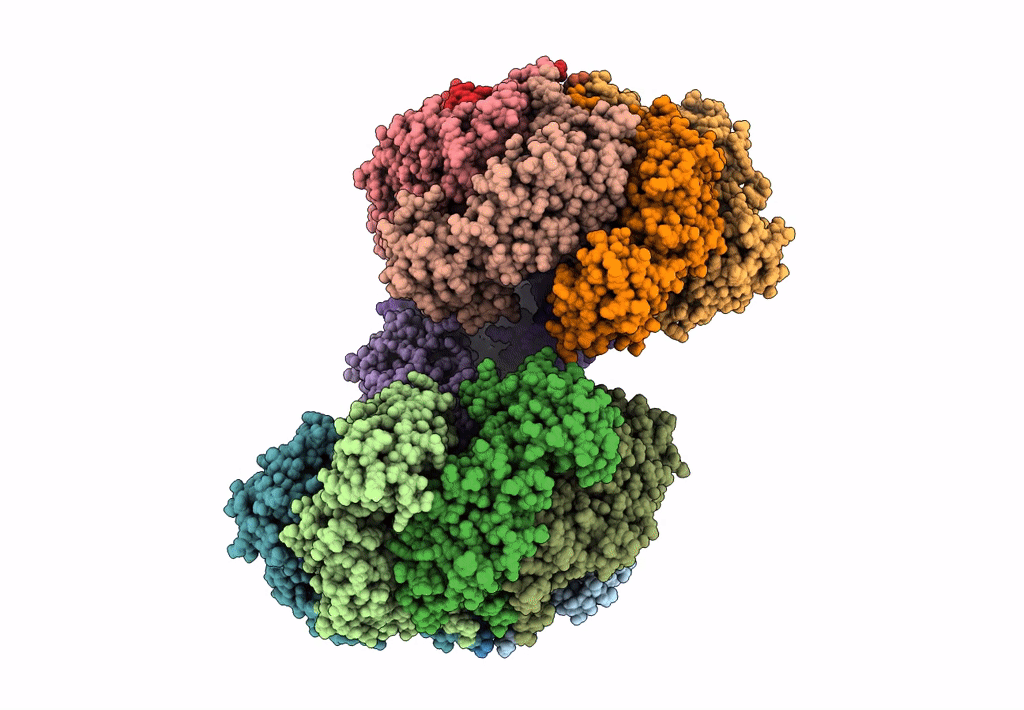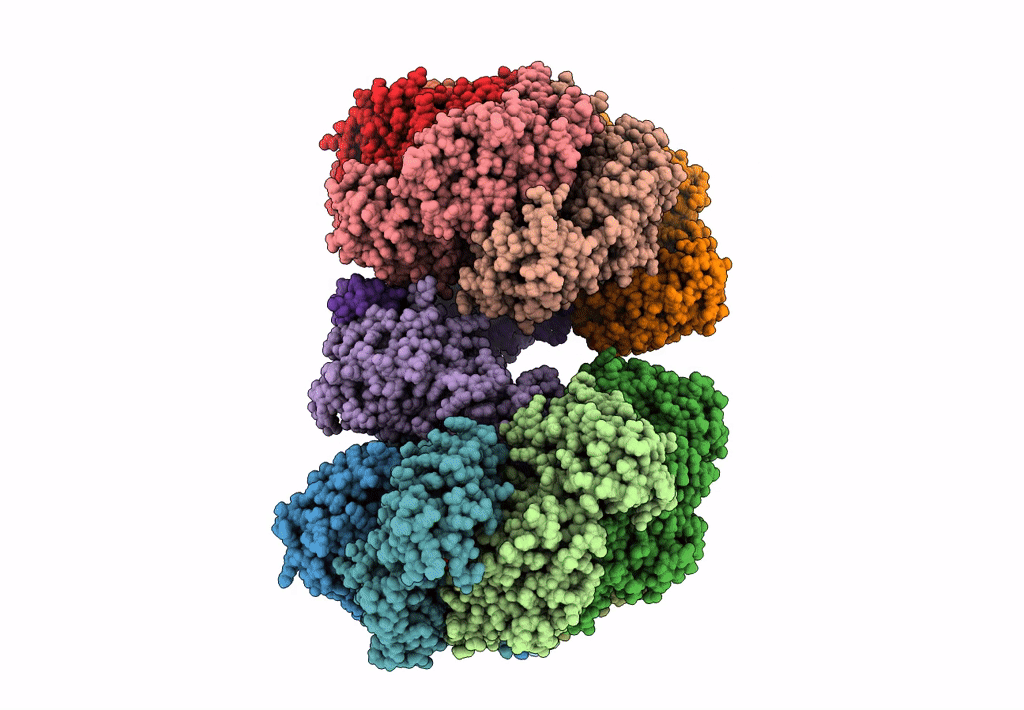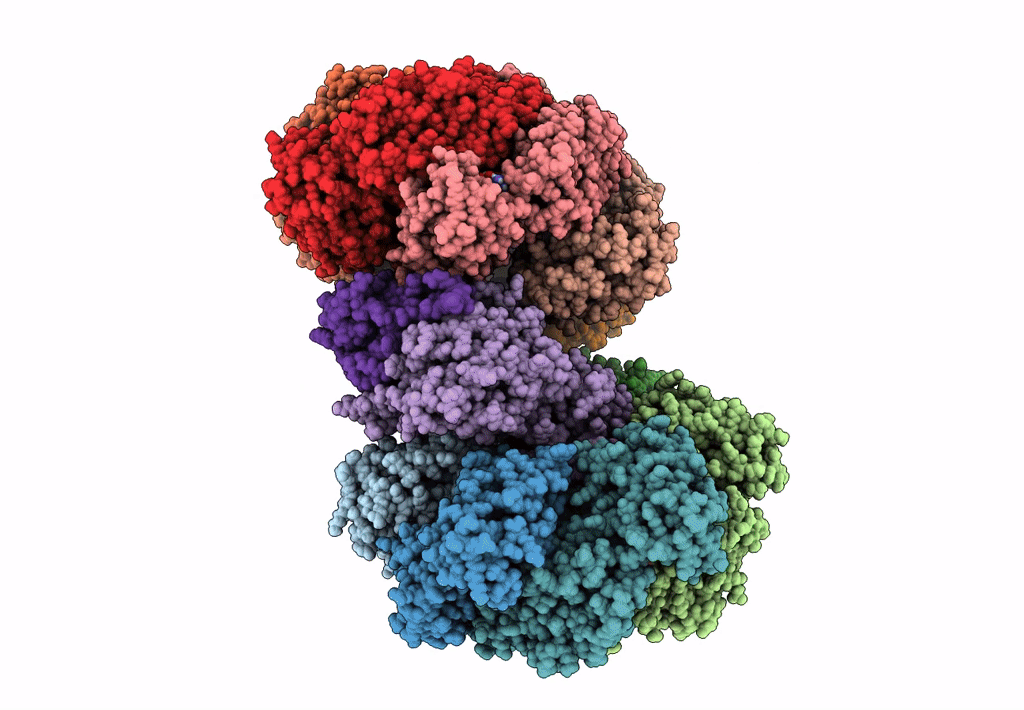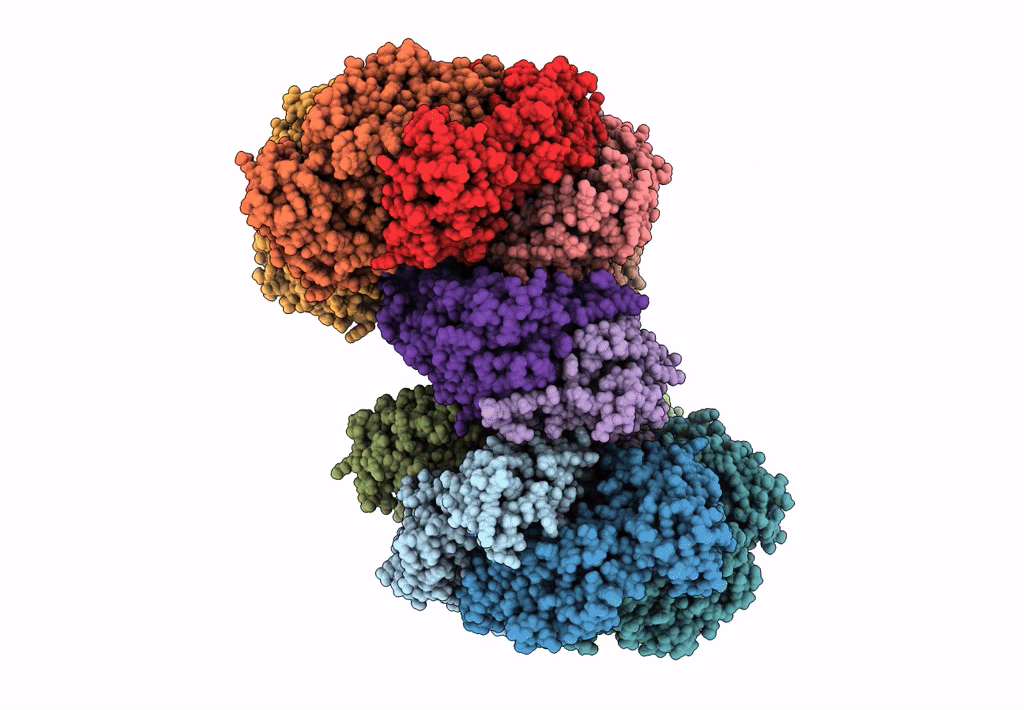 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: DNA BINDING PROTEIN Ligands: GNP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: V0S, CLR, NAG, NA, MPG |
 |
Structure Of Mod Subunit Of The Type Iii Restriction-Modification Enzyme Mbo45V
Organism: Mycoplasma bovis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-01-05 Classification: DNA BINDING PROTEIN Ligands: SFG, EDO |
 |
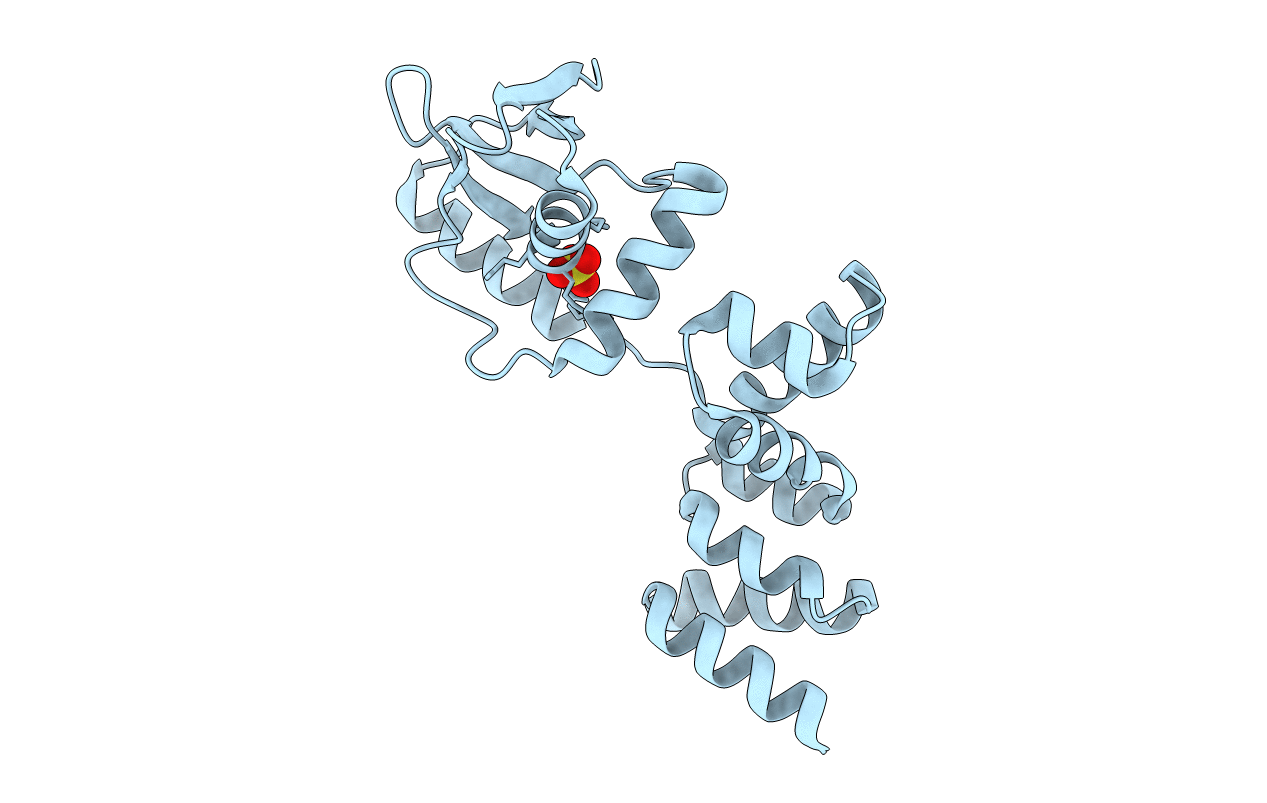
Crystal Structure Of C-Terminally Truncated Geobacillus Thermoleovorans Nucleoid Occlusion Protein Noc
Organism: Geobacillus thermoleovorans ccb_us3_uf5
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-02-17 Classification: DNA BINDING PROTEIN Ligands: SO4, GOL |
 |
Crystal Structure Of N- And C-Terminally Truncated Geobacillus Thermoleovorans Nucleoid Occlusion Protein Noc
Organism: Geobacillus thermoleovorans ccb_us3_uf5
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-02-17 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Staphylococcus aureus subsp. aureus usa300
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-01-20 Classification: DNA BINDING PROTEIN Ligands: SO4 |
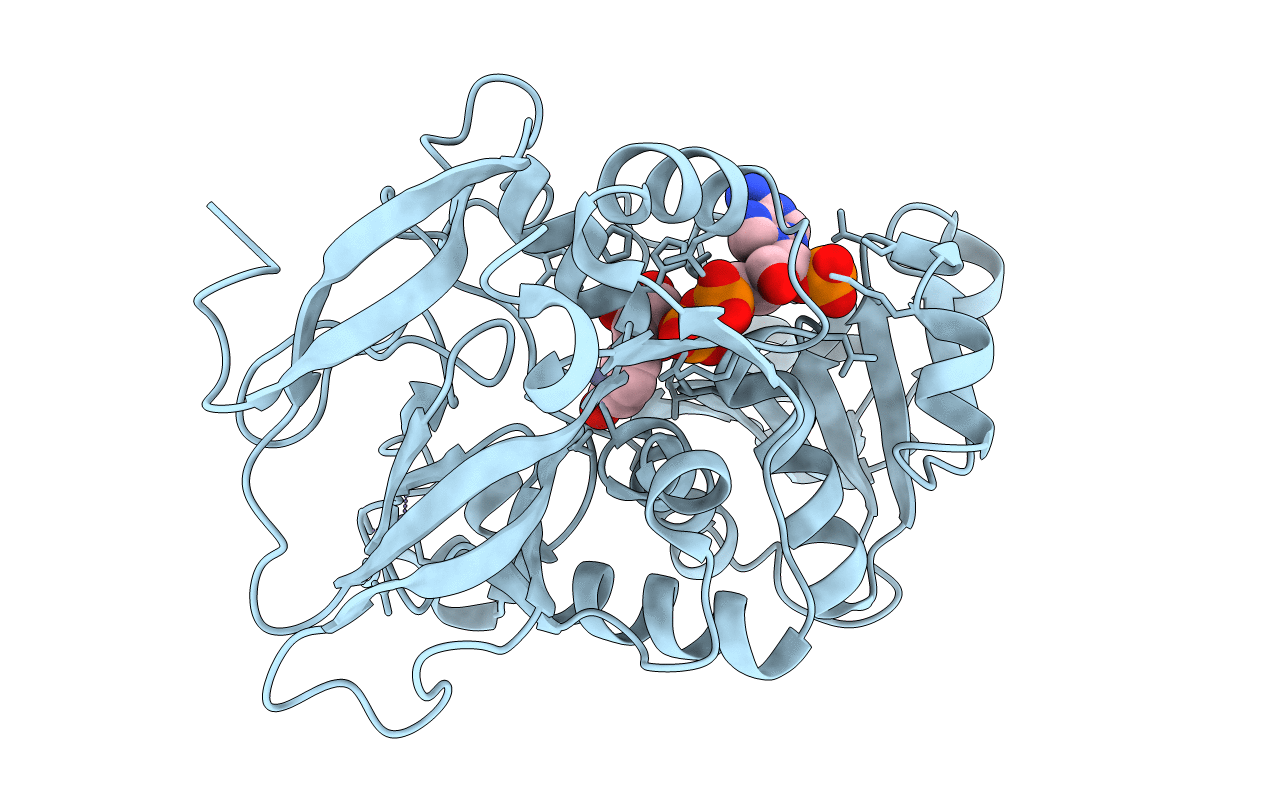 |
Crystal Structure Of 10-Hydroxygeraniol Dehydrogenase From Cantharanthus Roseus In Complex With Nadp+
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE Ligands: NAP, ZN |
 |
Crystal Structure Of 10-Hydroxygeraniol Dehydrogenase Apo Form From Cantharanthus Roseus
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:3.75 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Structure Of Mcrbc Without Dna Binding Domains (One Half Of The Full Complex)
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
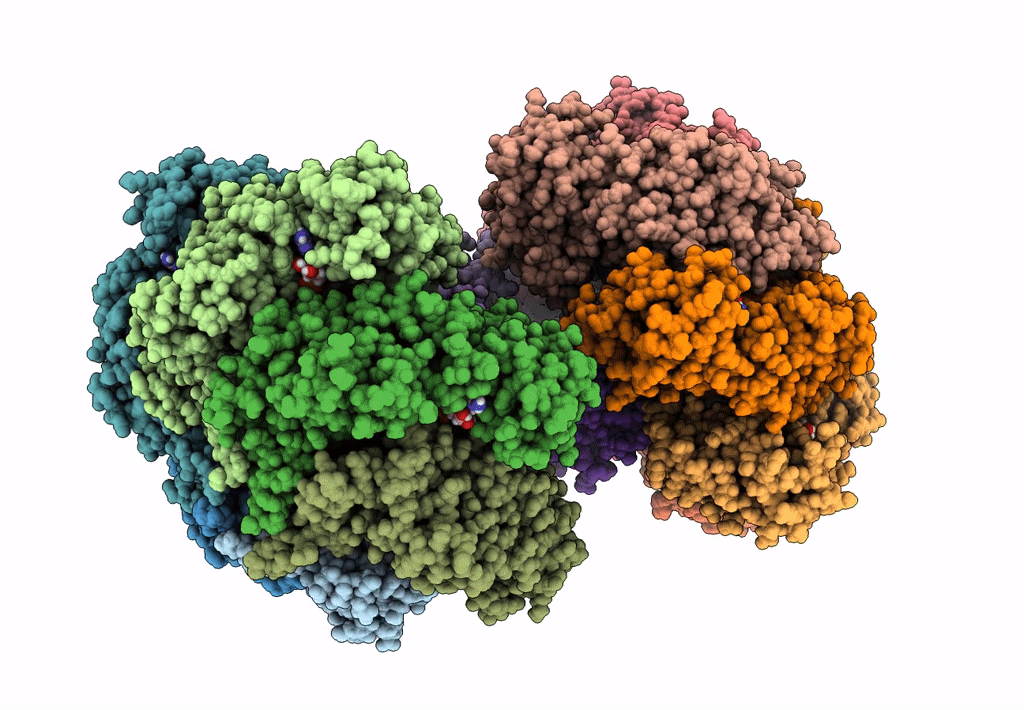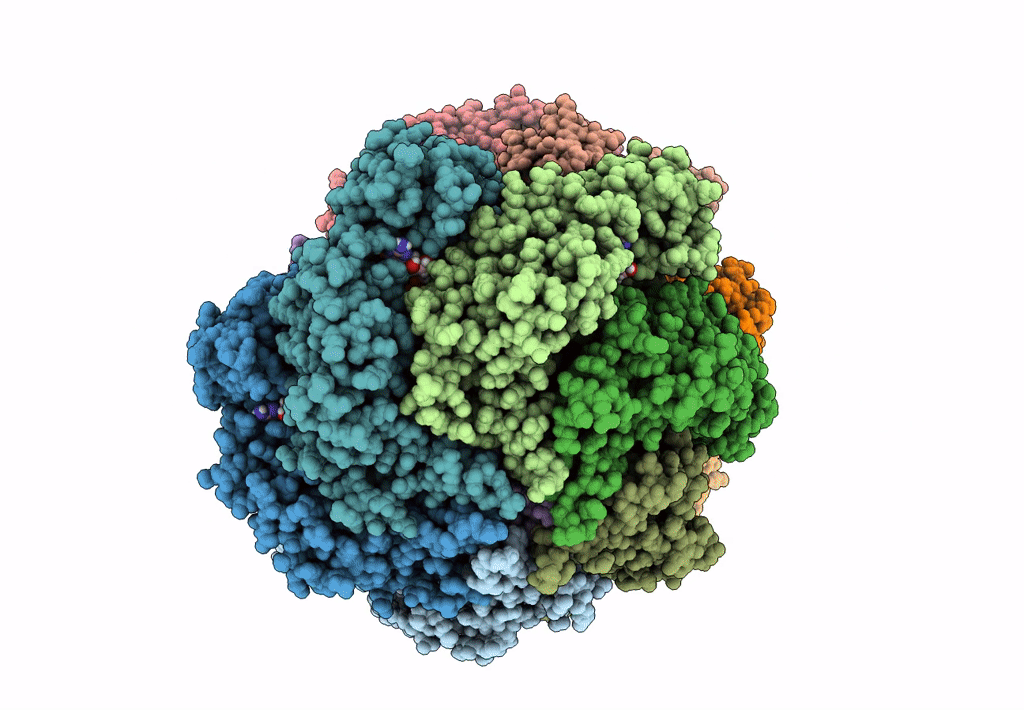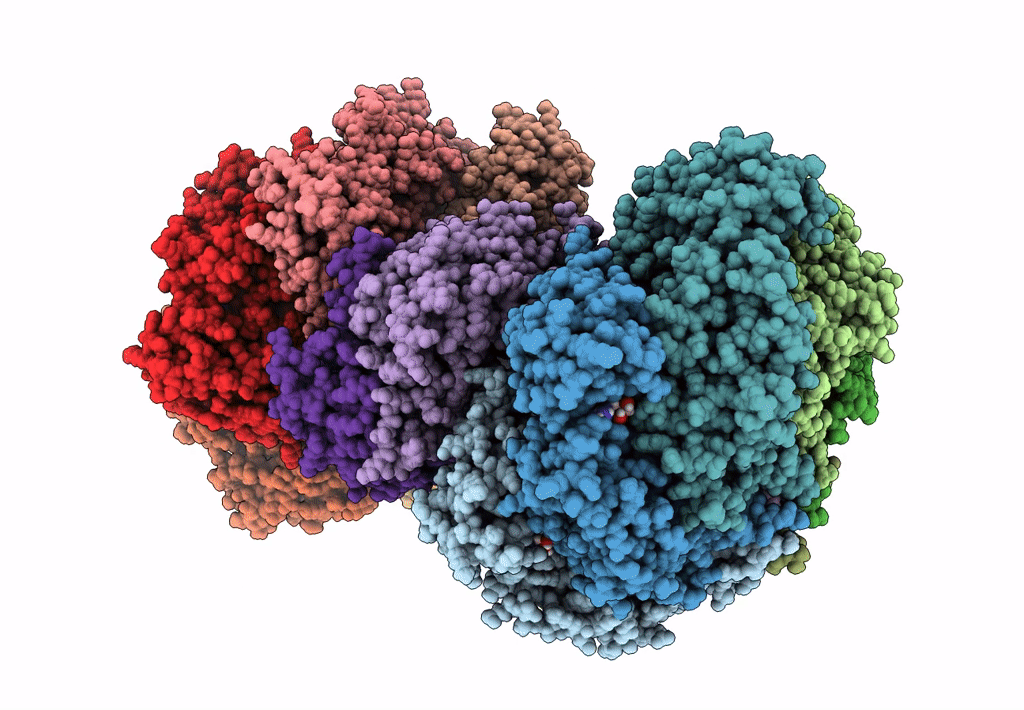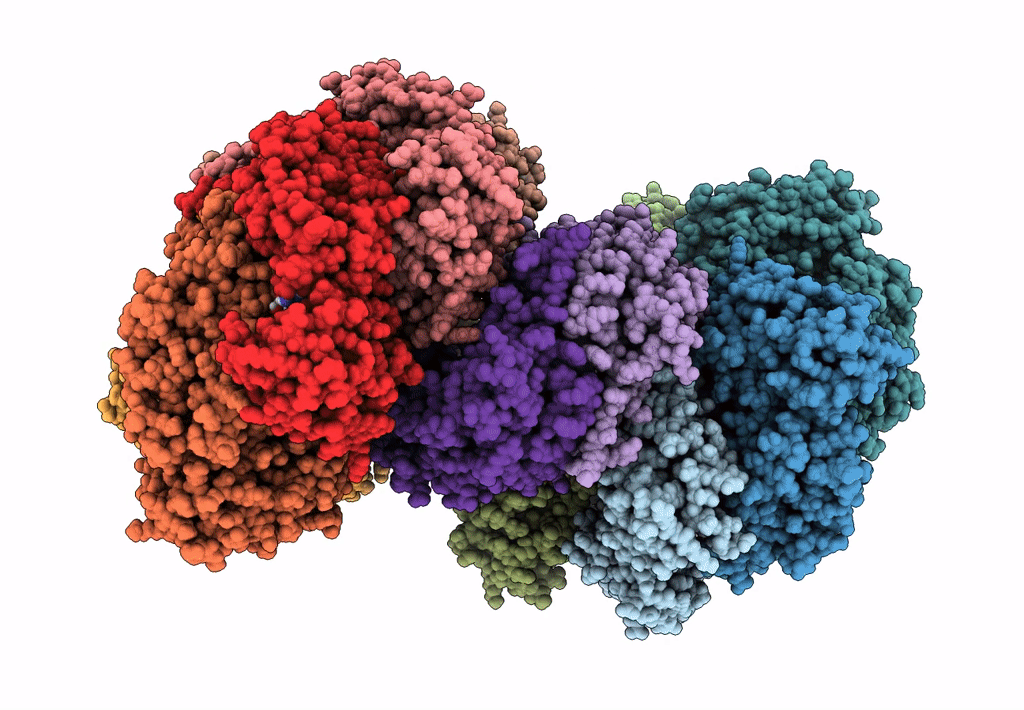
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
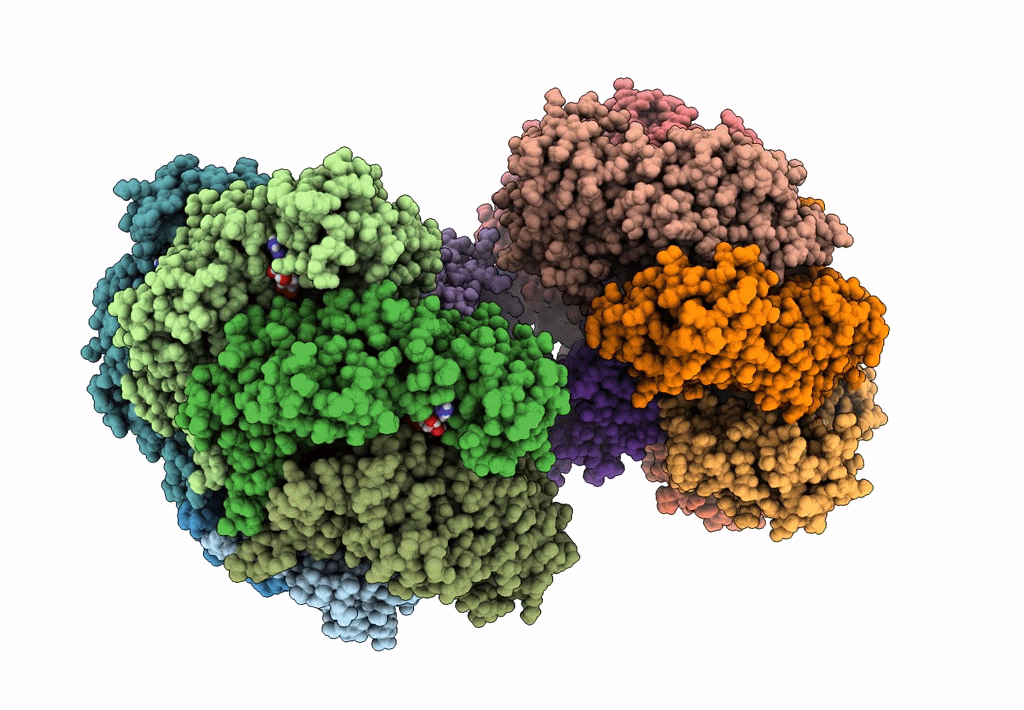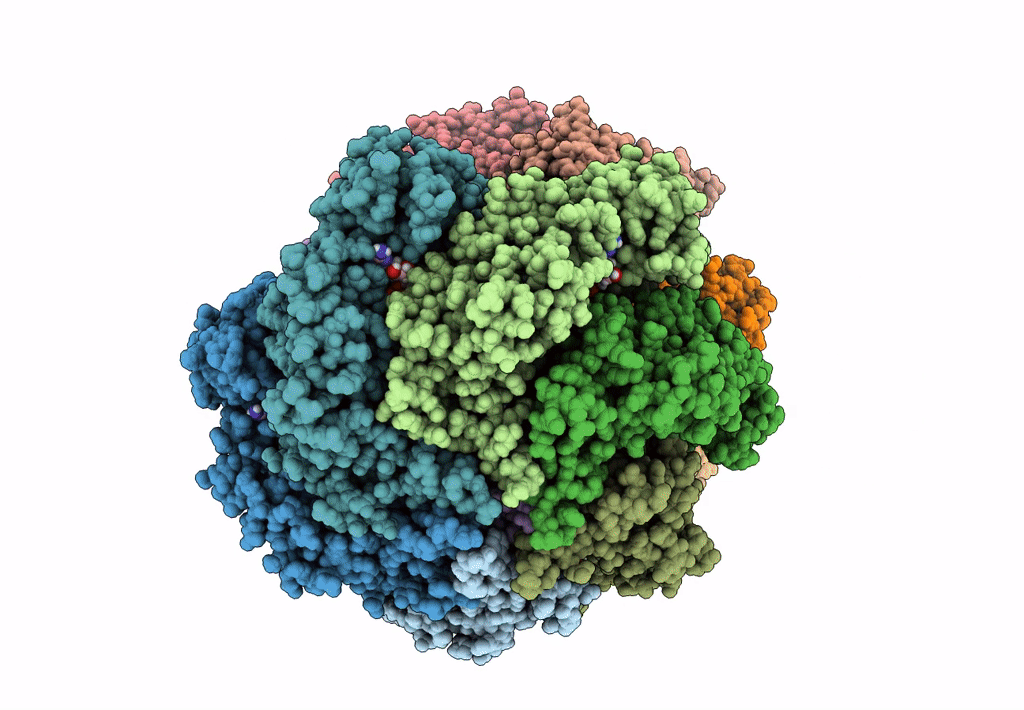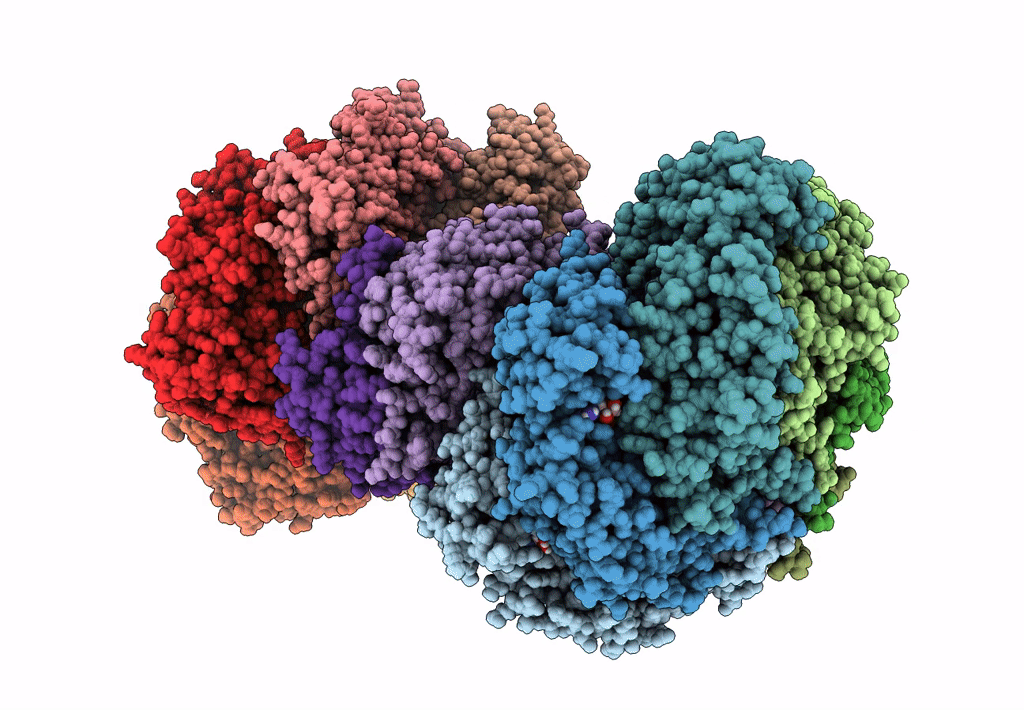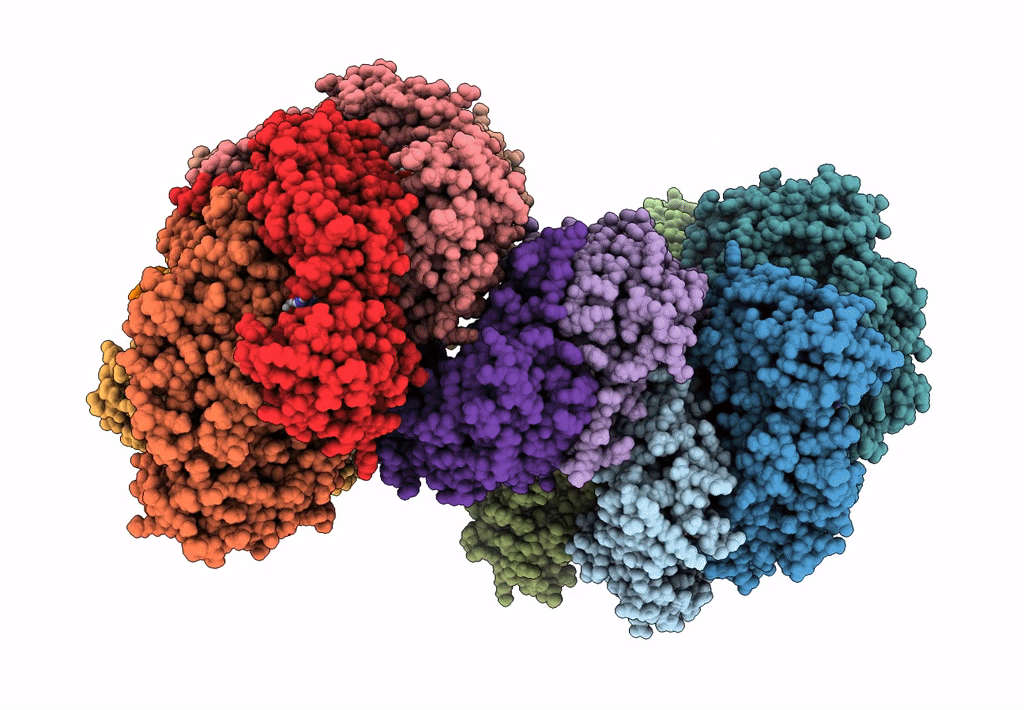 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |

