Search Count: 30
 |
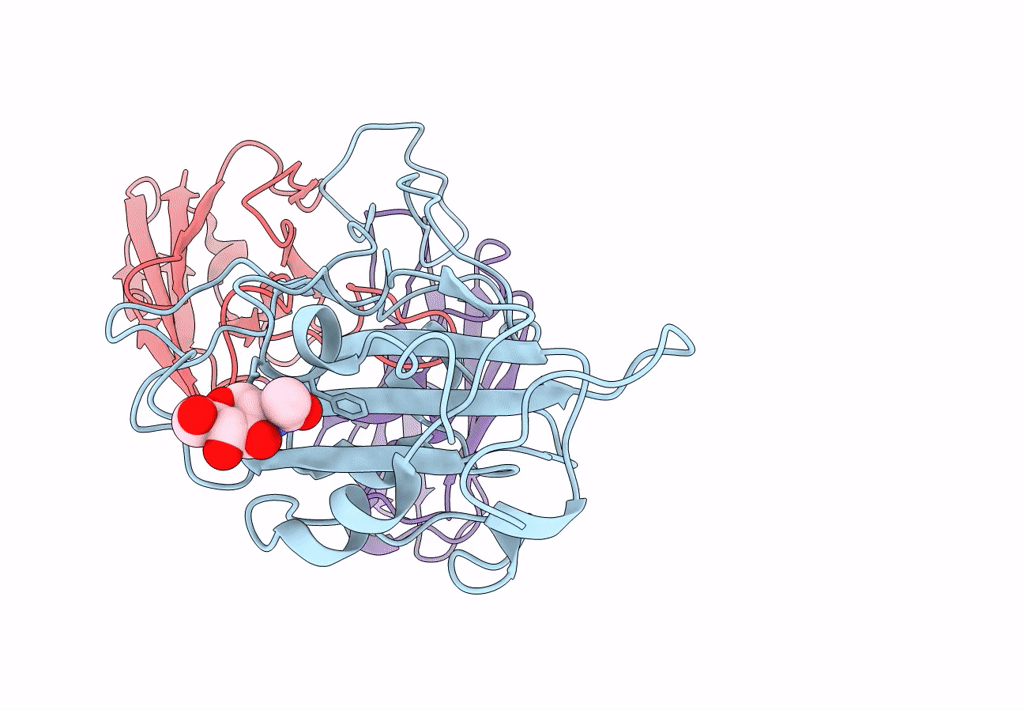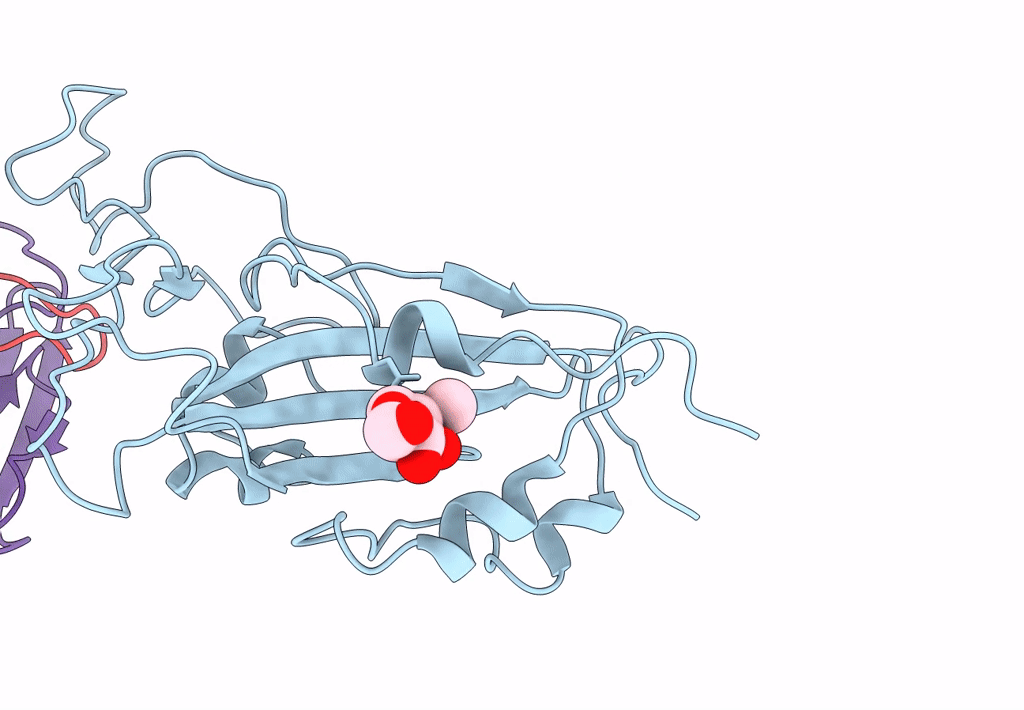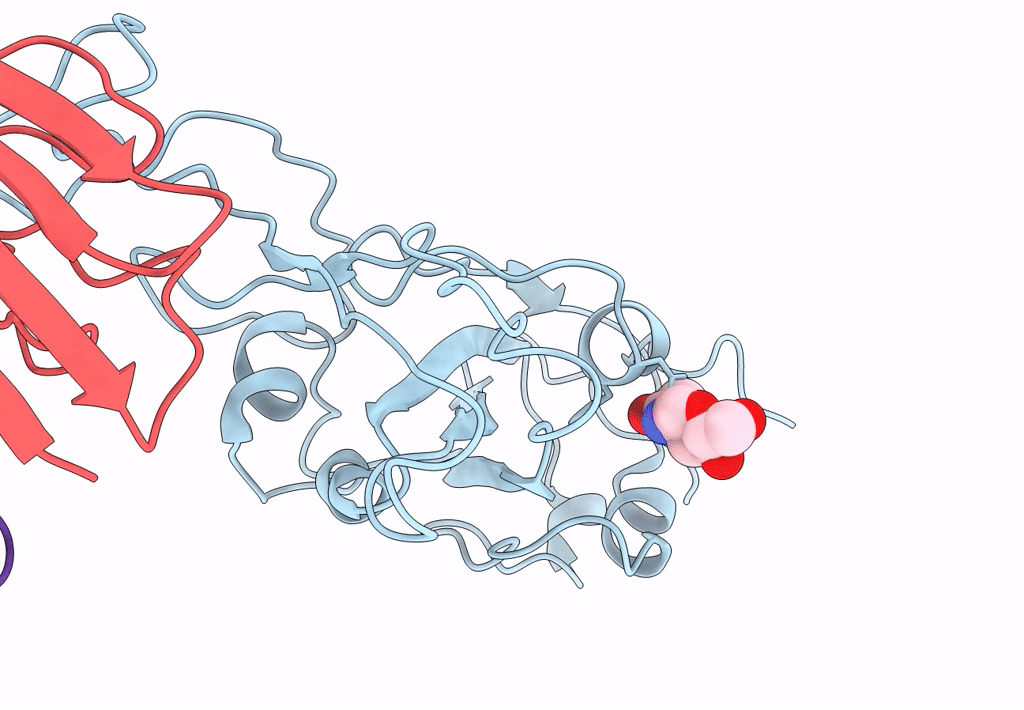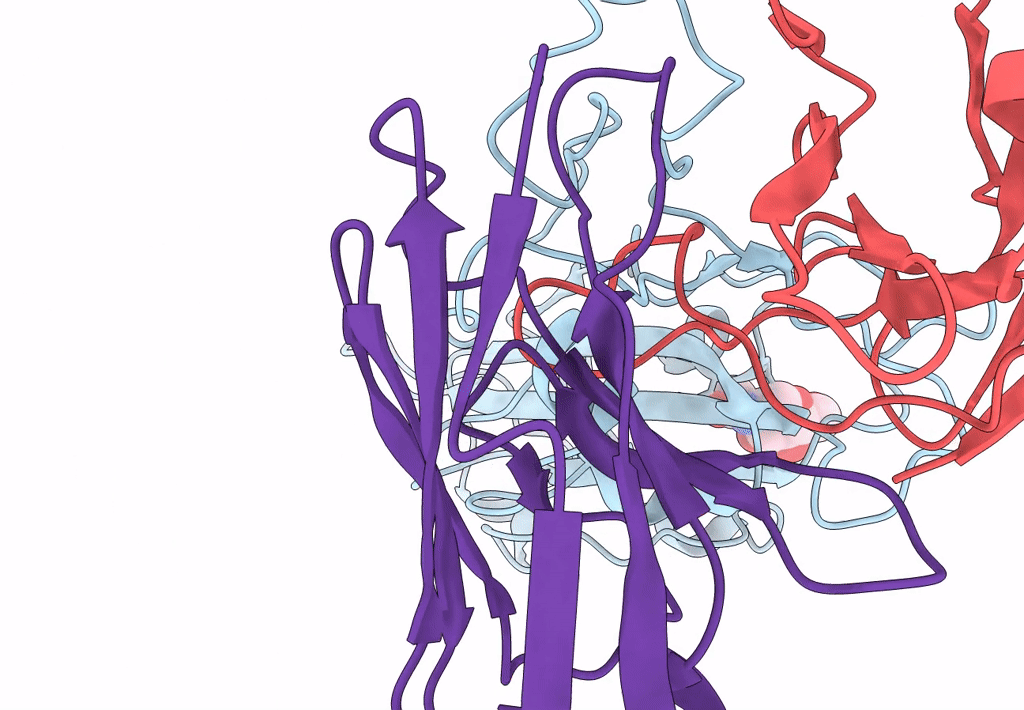
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab816
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab803
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
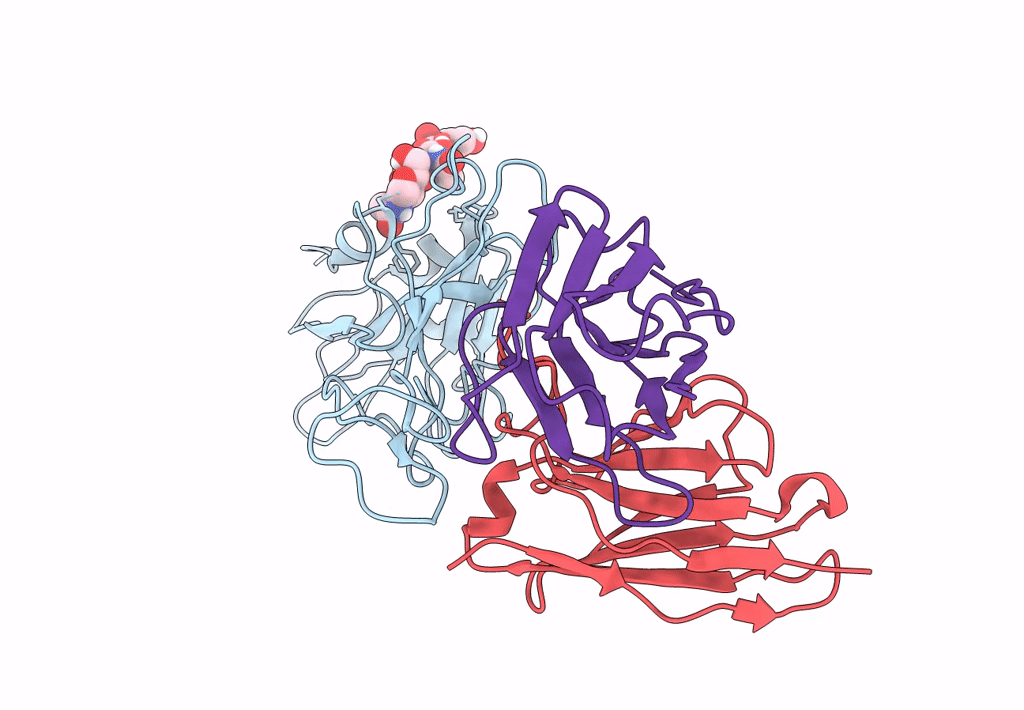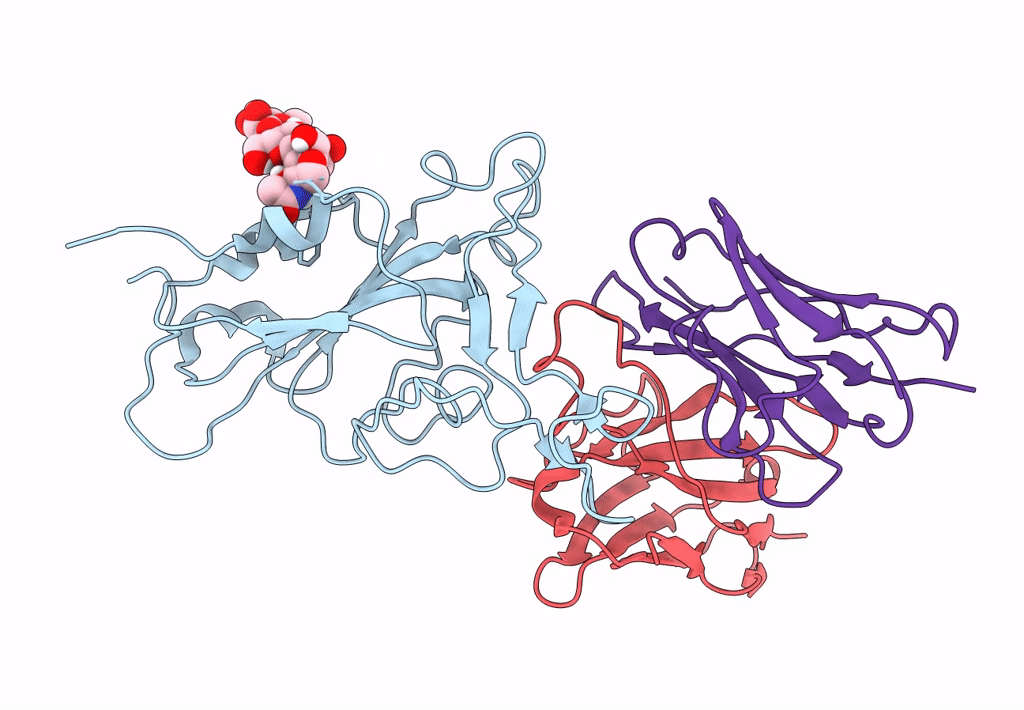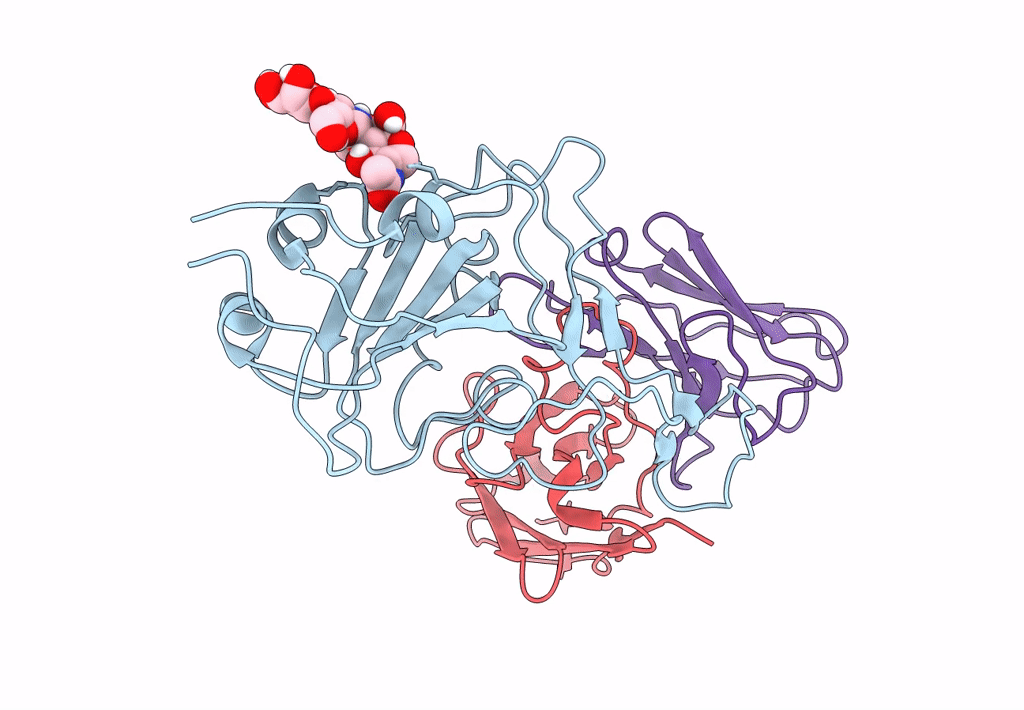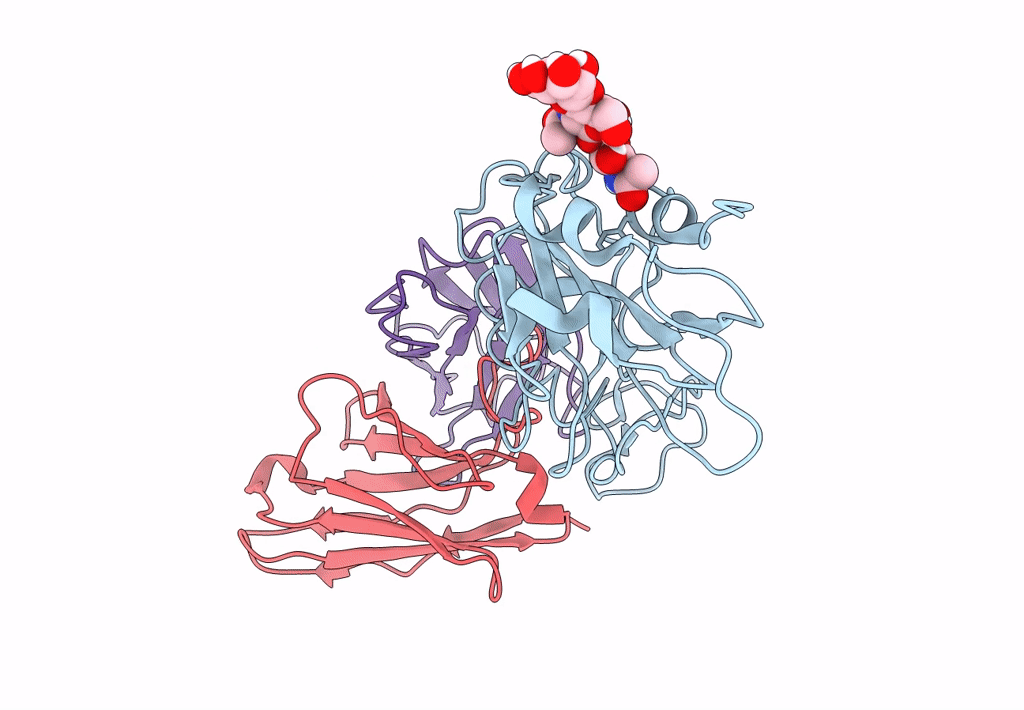
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab765
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
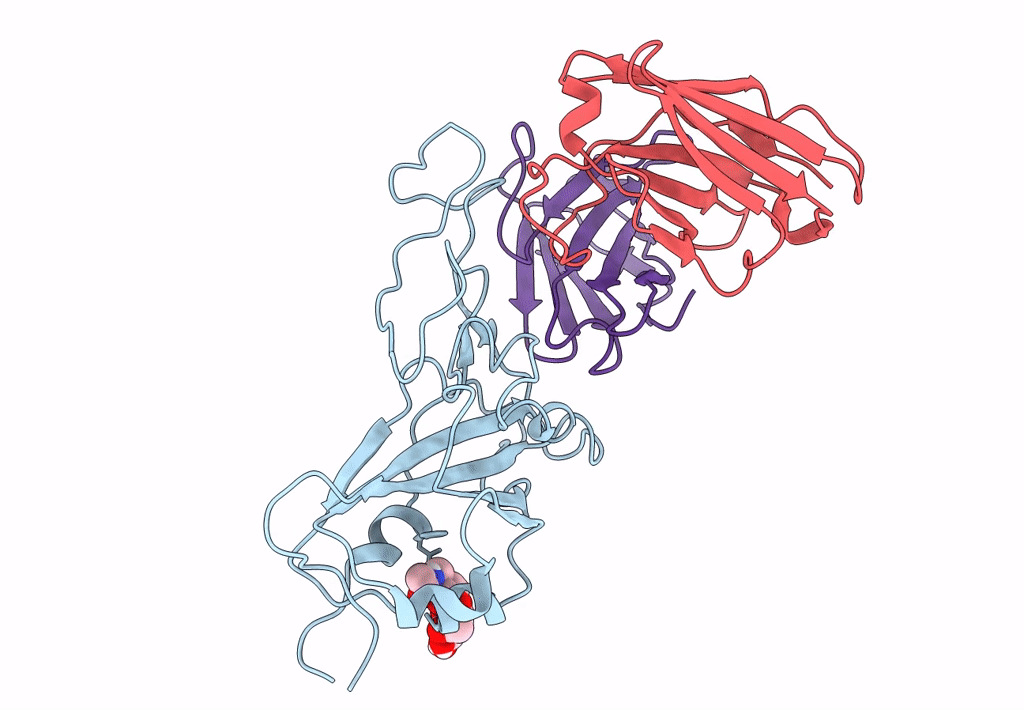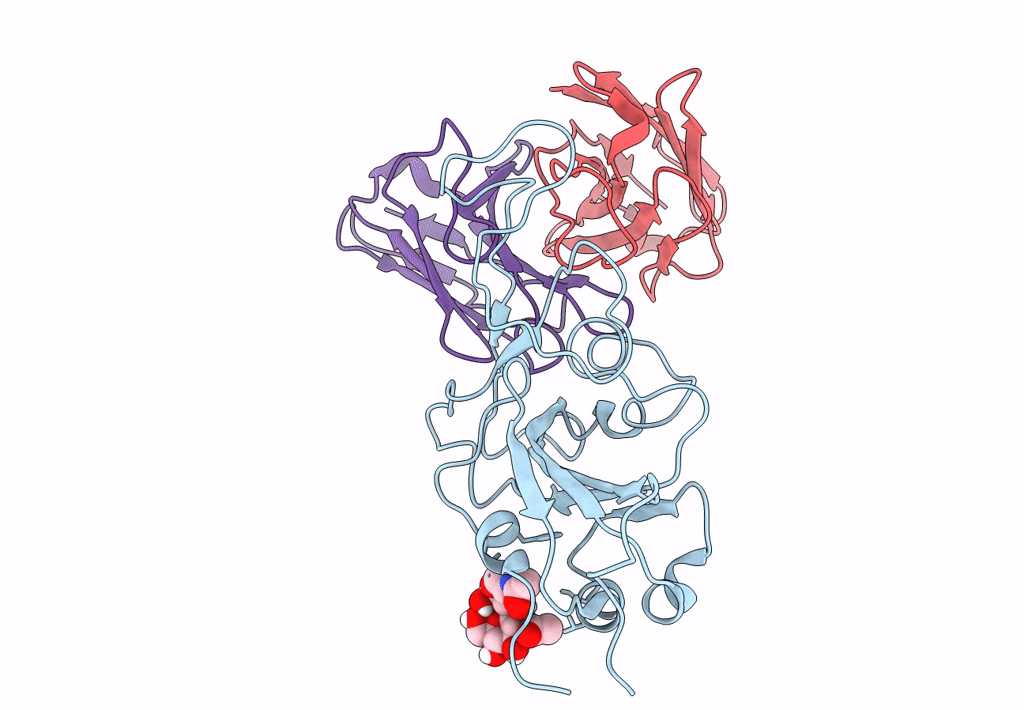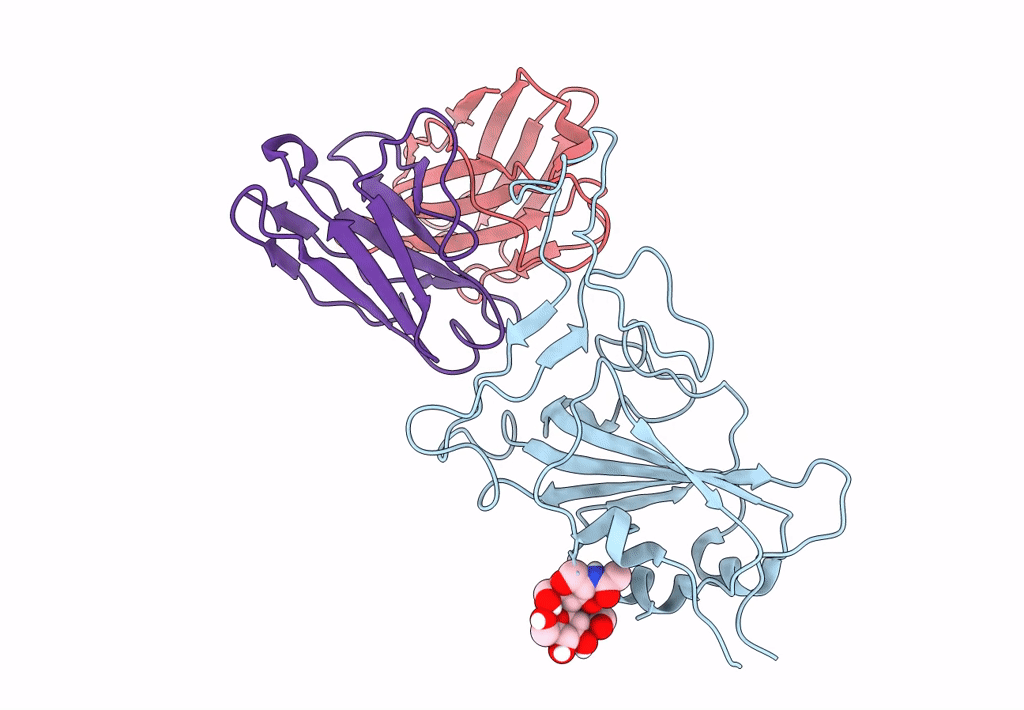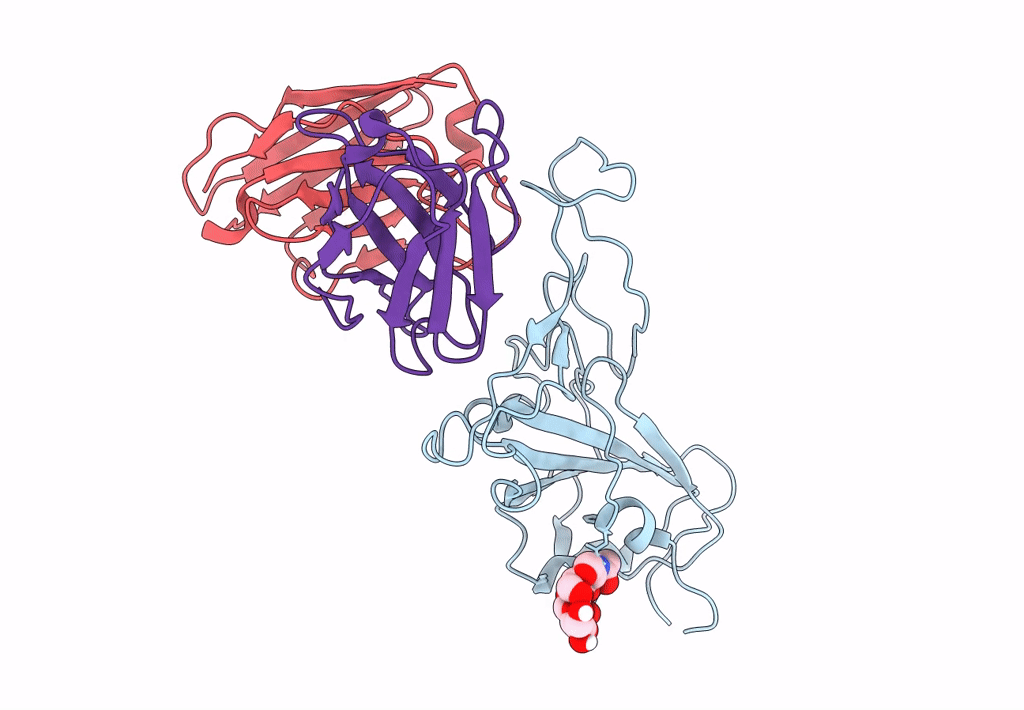
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab712
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab709
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN |
 |
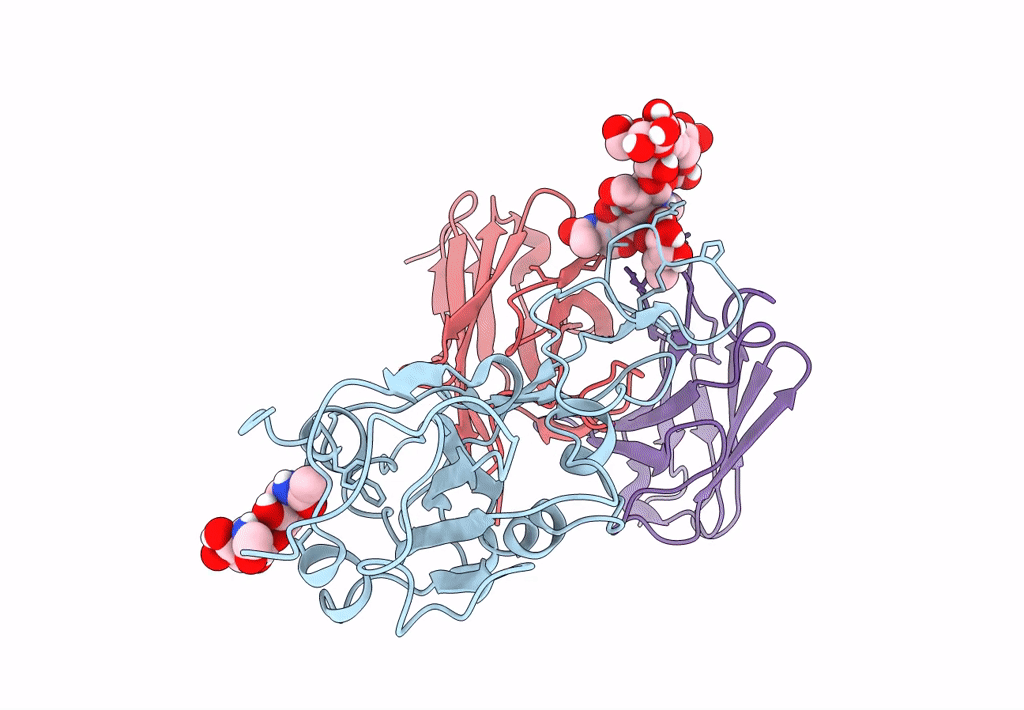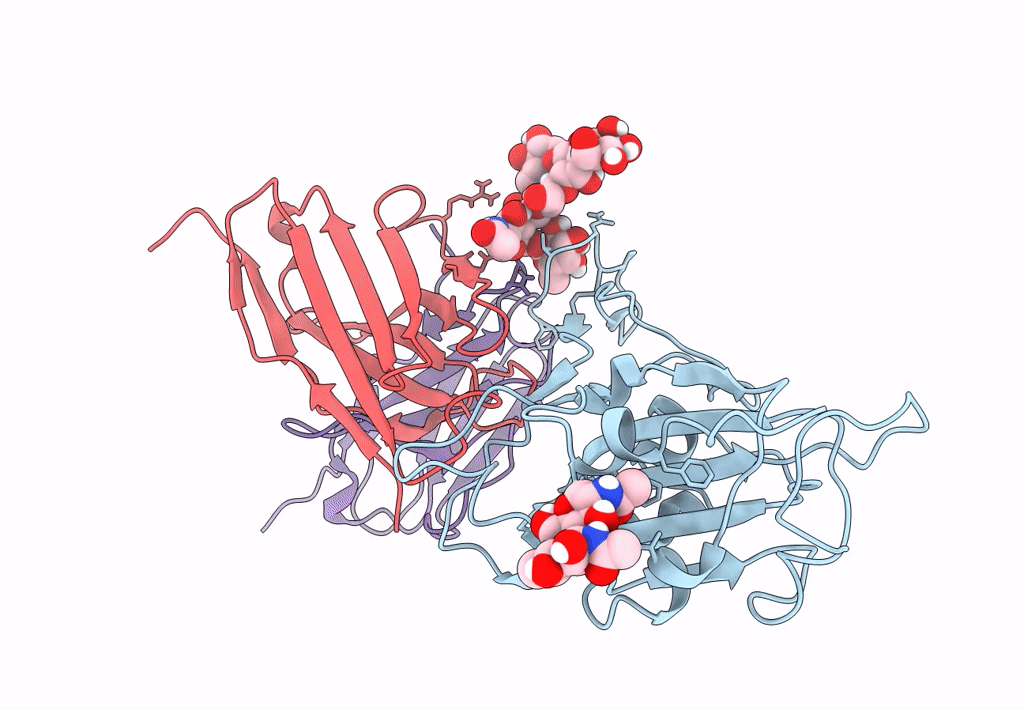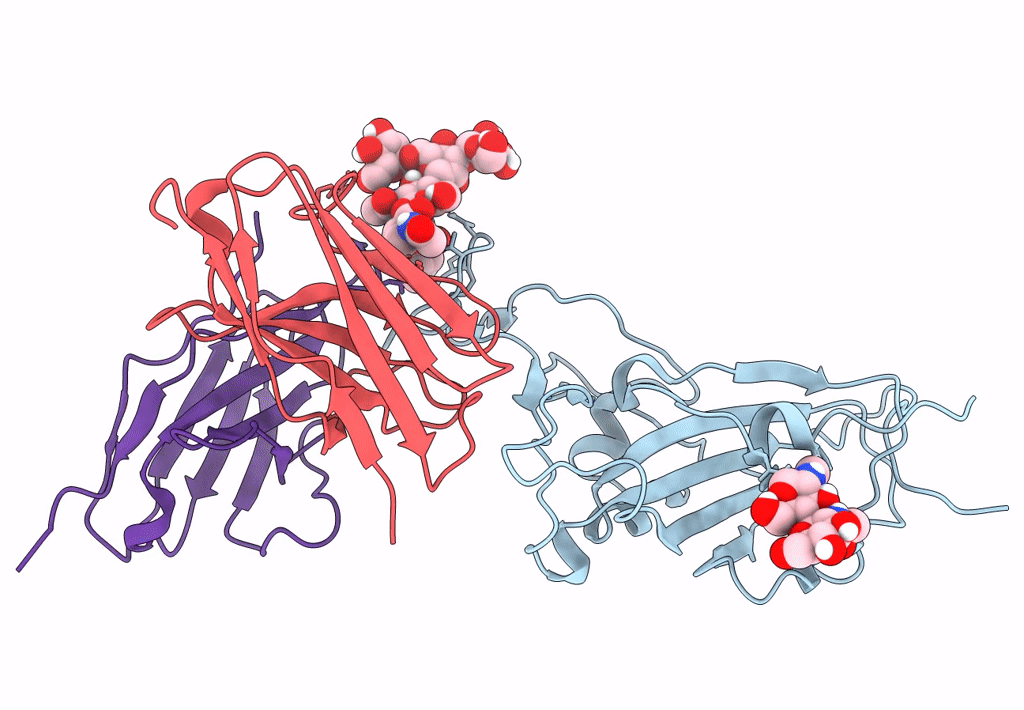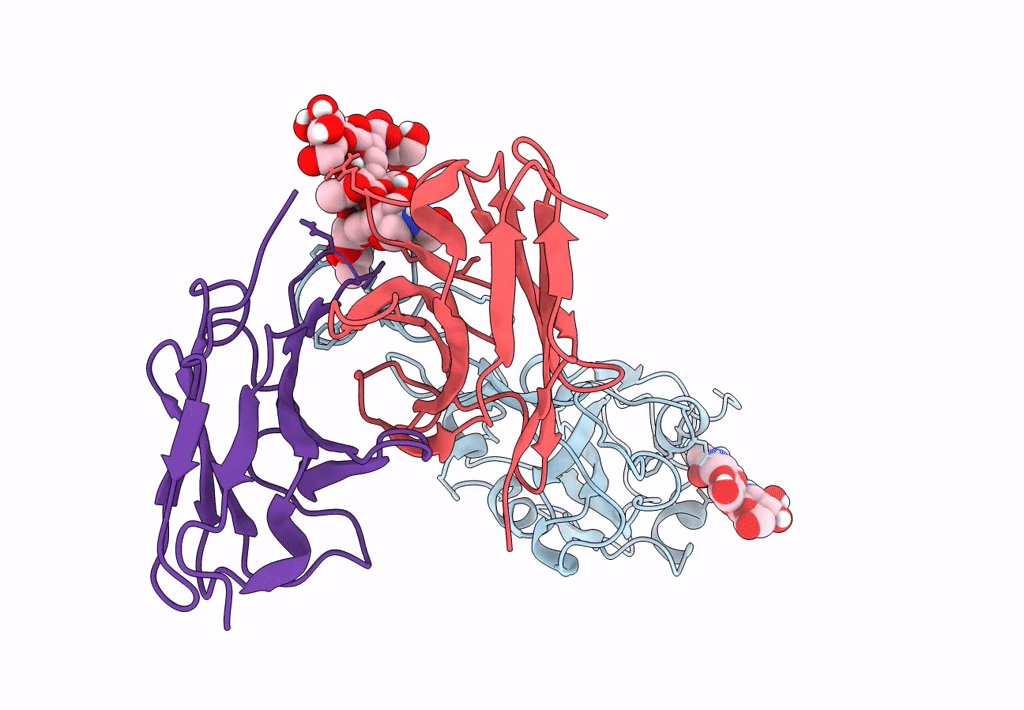
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab847
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab354
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab159
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
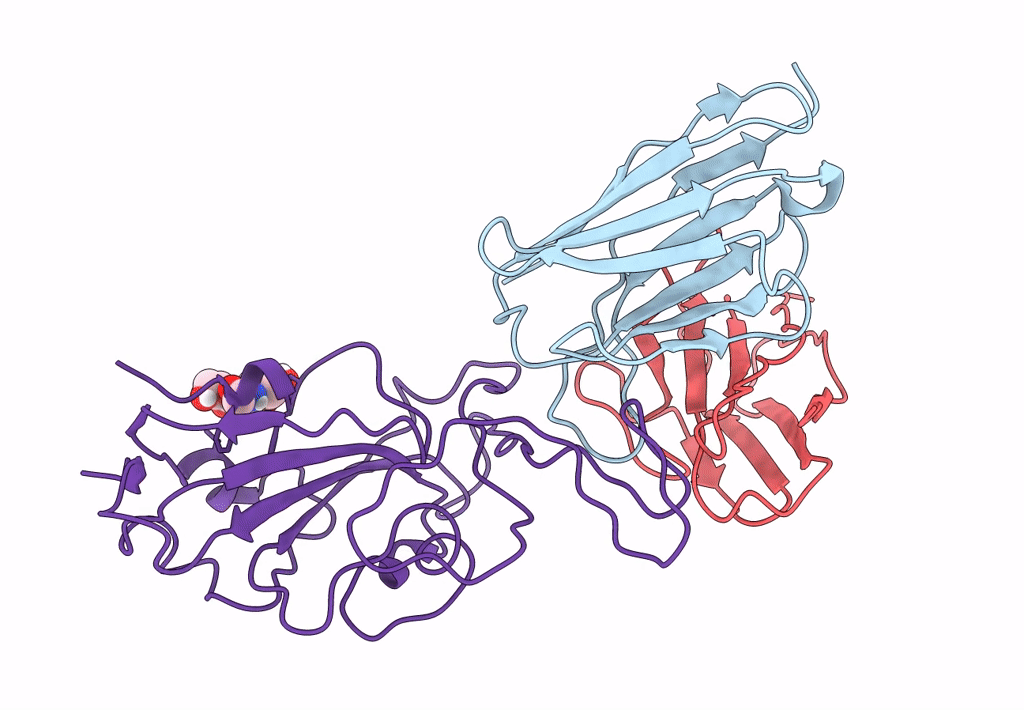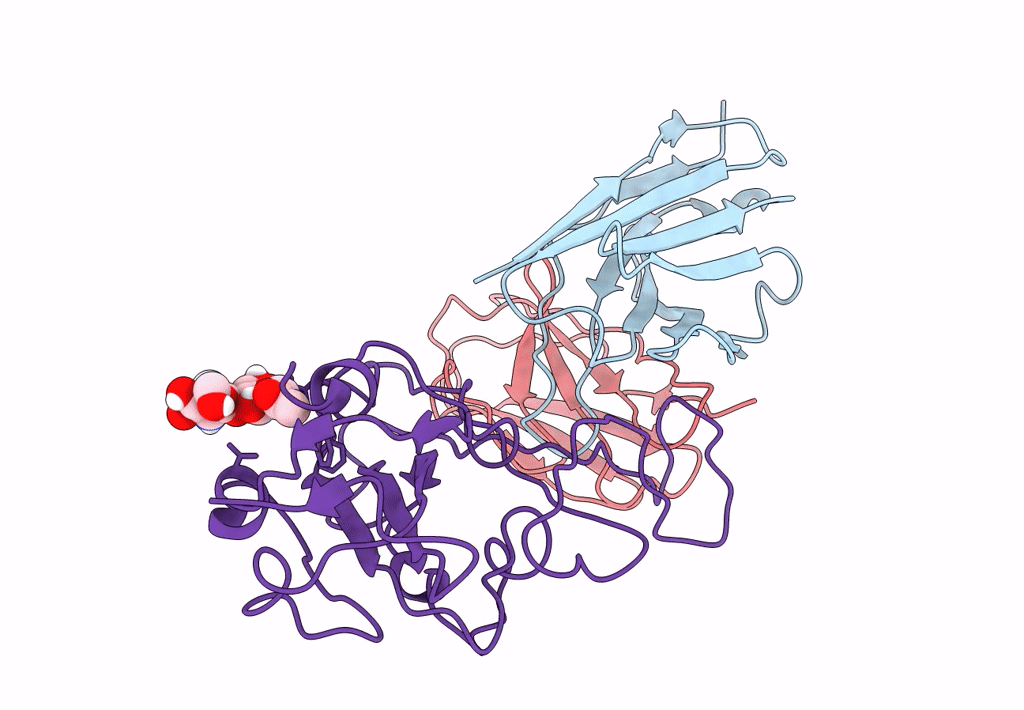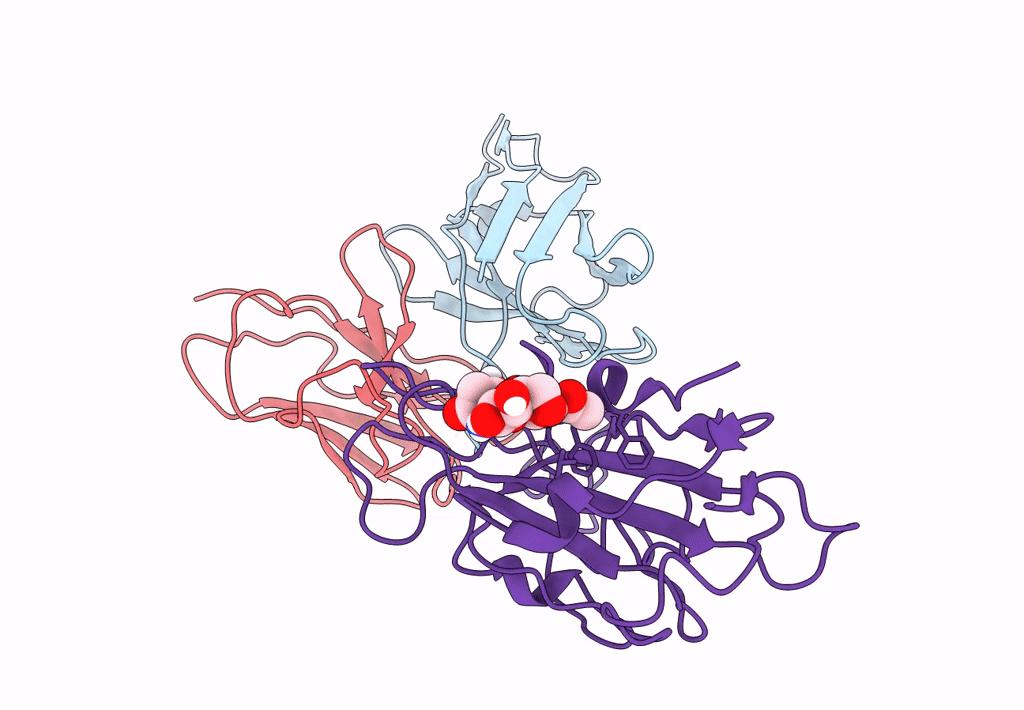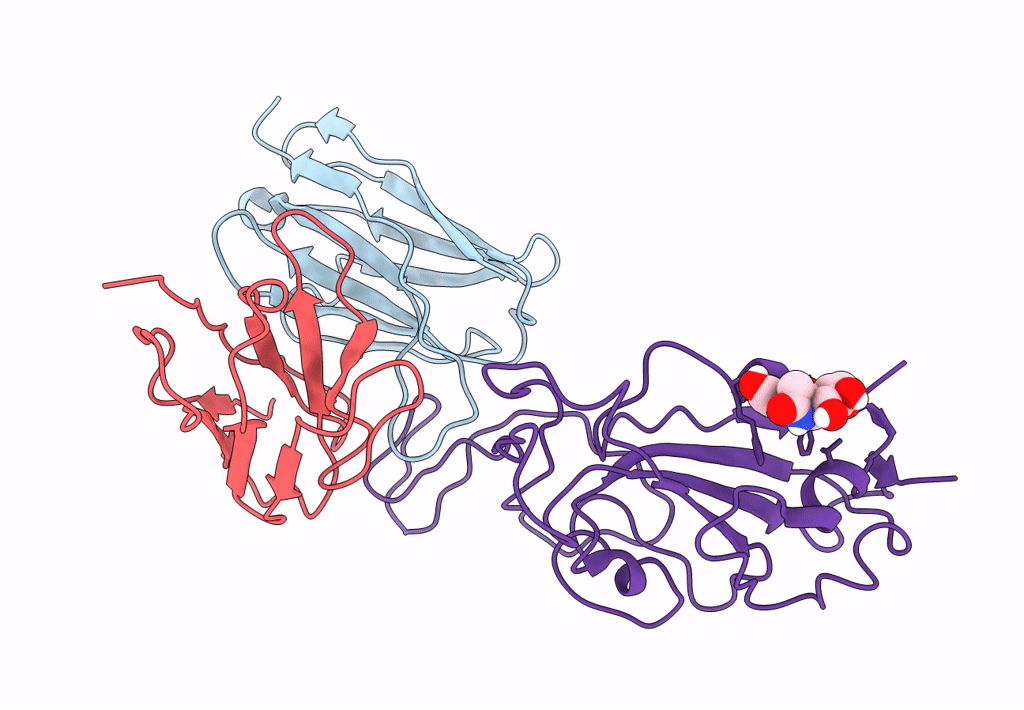
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab188
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab326
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With An Fv-Clasp Form Of A Human Neutralizing Antibody Ab496
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab445
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
Hemagglutinin Influenza A Virus (A/Okuda/1957(H2N2) Bound With A Neutralizing Antibody
Organism: Influenza a virus (a/okuda/1957(h2n2)), Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of The Bacillus Subtilis Smc Head Domain Complexed With The Cognate Scpa C-Terminal Domain
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-03-15 Classification: DNA BINDING PROTEIN/CELL CYCLE |
 |
Crystal Structure Of The Bacillus Subtilis Smc Head Domain Complexed With The Cognate Scpa C-Terminal Domain And Soaked Atp
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2017-03-15 Classification: DNA BINDING PROTEIN/CELL CYCLE Ligands: ATP |
 |
Crystal Structure Of An Engaged Dimer Of The Geobacillus Stearothermophilus Smc Head Domain
Organism: Geobacillus stearothermophilus 10
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-03-15 Classification: DNA BINDING PROTEIN, CELL CYCLE Ligands: AGS, MG |
 |
Crystal Structure Of An Asymmetric Dimer Of The Geobacillus Stearothermophilus Smc Hinge Domain
Organism: Geobacillus stearothermophilus 10
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-15 Classification: DNA BINDING PROTEIN, CELL CYCLE |
 |
Organism: Escherichia coli, Synthetic construct, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-04-24 Classification: CELL CYCLE |
 |
Crystal Structure Of Dimer Of Scpb N-Terminal Domain Complexed With Scpa Peptide
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2013-04-24 Classification: CELL CYCLE |

