Search Count: 22
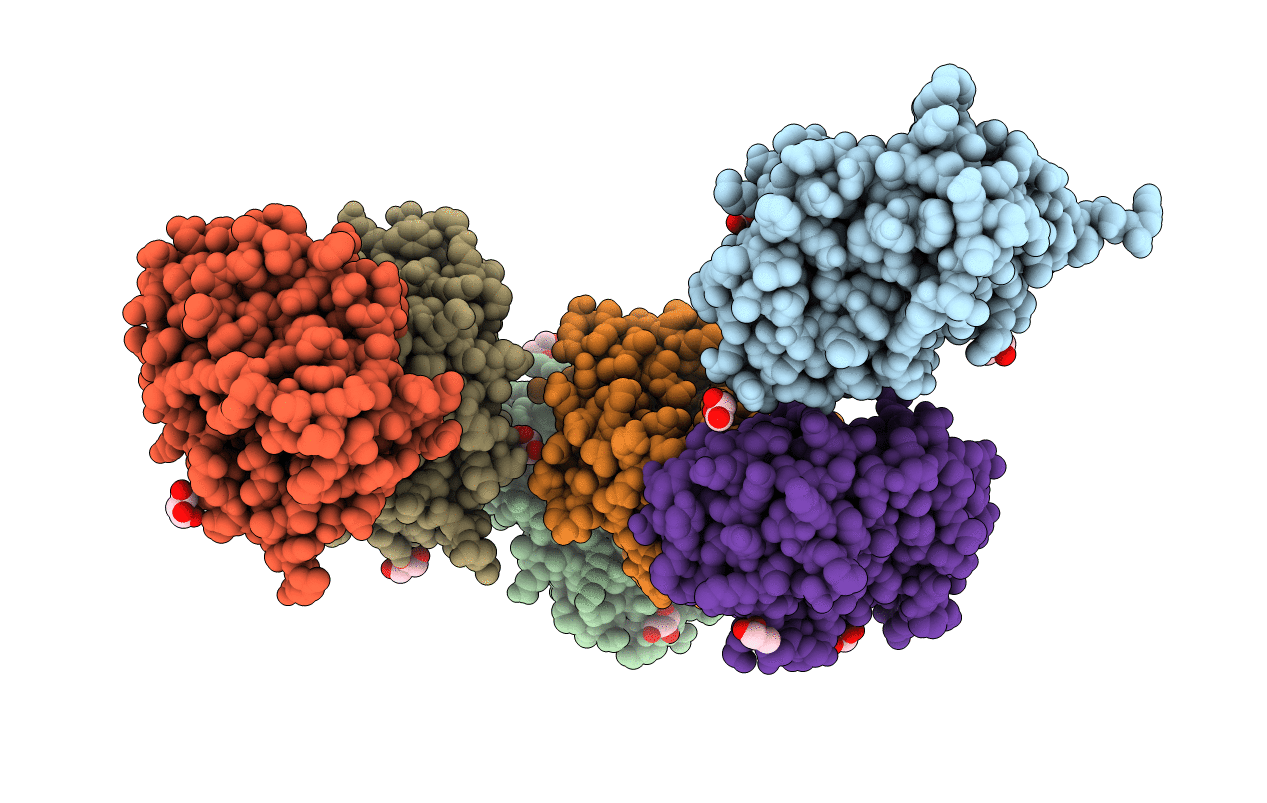 |
Crystal Structure Of Bh1352 2-Deoxyribose-5-Phosphate From Bacillus Halodurans, K184L Mutant
Organism: Bacillus halodurans c-125
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2019-10-23 Classification: LYASE Ligands: GOL |
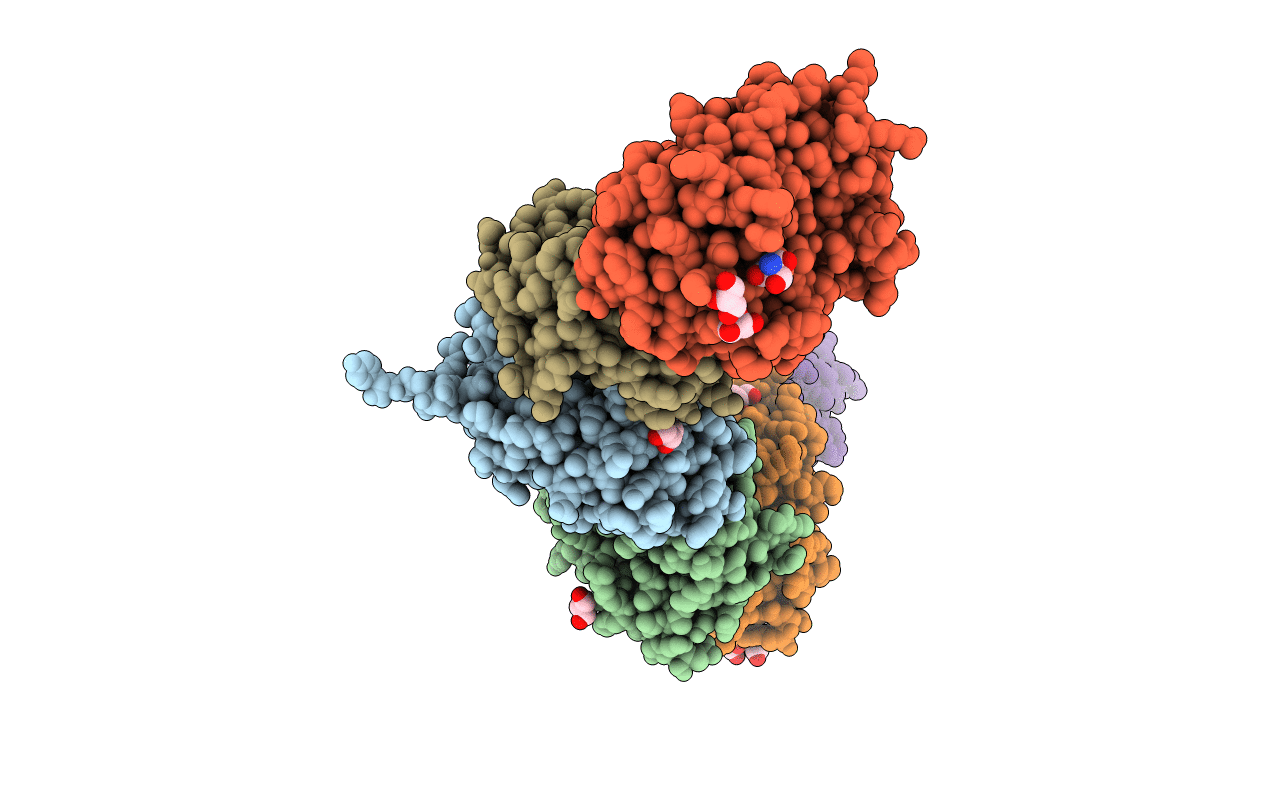 |
Crystal Structure Of Bh1352 2-Deoxyribose-5-Phosphate From Bacillus Halodurans
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-10-16 Classification: TRANSFERASE Ligands: GOL, TRS |
 |
Human Peroxisome Proliferator-Activated Receptor (Ppar) Delta In Complexed With A Potent And Selective Agonist
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-01 Classification: PROTEIN BINDING Ligands: 8RR, K, CL, PGO, JZR |
 |
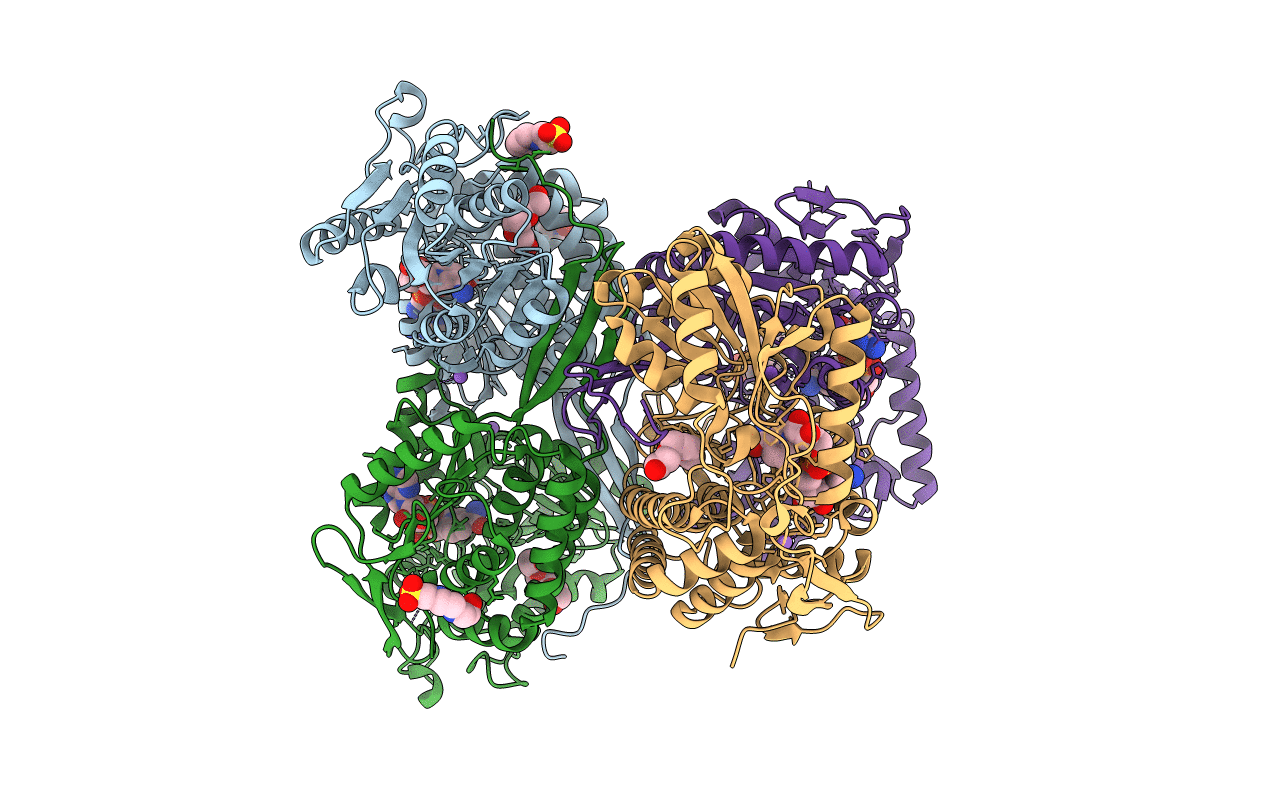
X-Ray Crystal Structure Of A Novel Aldo-Keto Reductases For The Biocatalytic Conversion Of 3-Hydroxybutanal To 1,3-Butanediol
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-02-15 Classification: TRANSLATION Ligands: SO4, CL, NDP |
 |
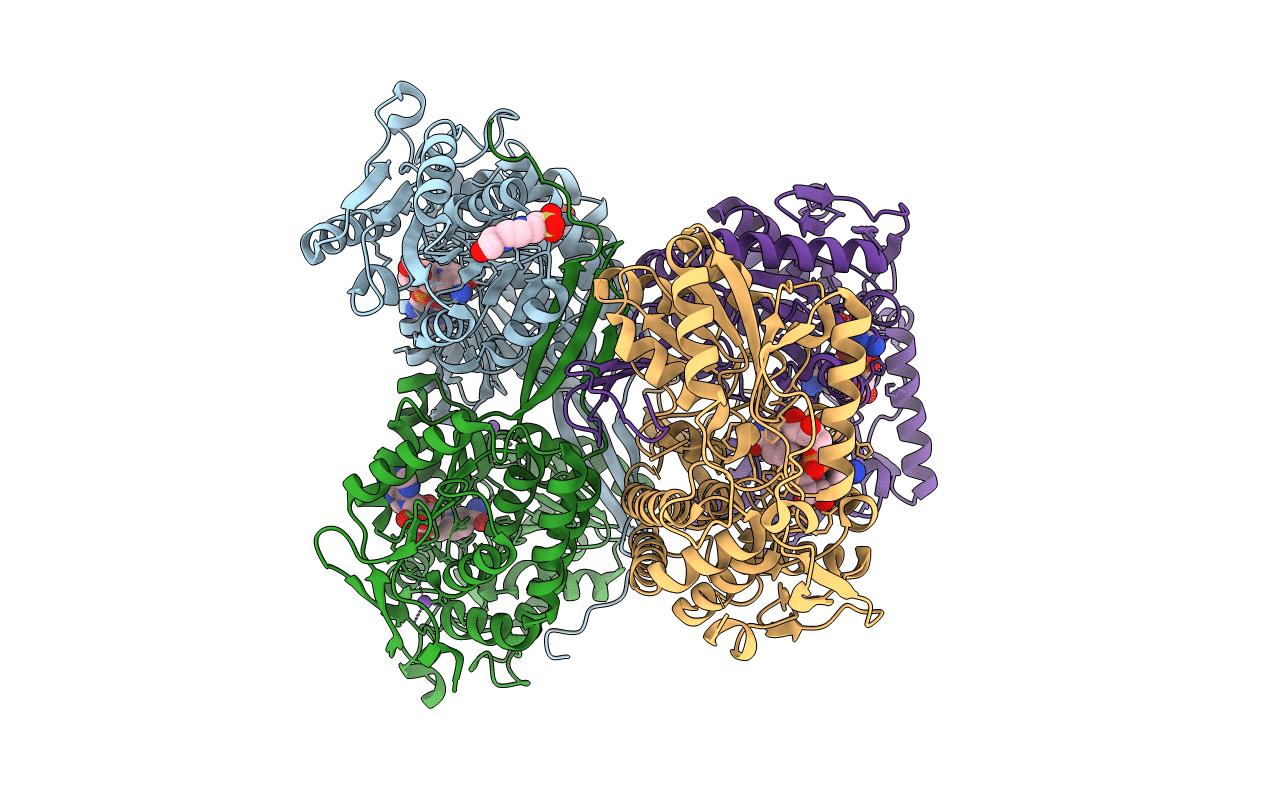
1.72 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) P449M Point Mutant From Staphylococcus Aureus In Complex With Nad+ And Bme-Modified Cys289
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE Ligands: NAD, NA, EPE, PGE |
 |
2.11 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) P449M/Y450L Double Mutant From Staphylococcus Aureus In Complex With Nad+ And Bme-Modified Cys289
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE Ligands: NAD, NA, PGE, EPE |
 |
2.25 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) Y450L Point Mutant From Staphylococcus Aureus In Complex With Nad+ And Bme-Modified Cys289
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-10-14 Classification: OXIDOREDUCTASE Ligands: NAD, NA, EPE |
 |
2.85 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) H448F/P449M Double Mutant From Staphylococcus Aureus In Complex With Nad+ And Bme-Free Cys289
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-06-17 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
2.60 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) H448F/Y450L Double Mutant From Staphylococcus Aureus In Complex With Nad+ And Bme-Free Cys289
Organism: Staphylococcus aureus (strain col)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-05-27 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
1.65 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus With Bme-Modified Cys289 And Peg Molecule In Active Site
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE |
 |
2.6 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) G234S Mutant From Staphylococcus Aureus (Idp00699) In Complex With Nad+ And Bme-Free Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-07-02 Classification: OXIDOREDUCTASE Ligands: NAD, ACT |
 |
1.85 Angstrom Resolution Crystal Structure Of Apo Betaine Aldehyde Dehydrogenase (Betb) G234S Mutant From Staphylococcus Aureus (Idp00699) With Bme-Free Sulfinic Acid Form Of Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-06-18 Classification: OXIDOREDUCTASE |
 |
1.90 Angstrom Resolution Crystal Structure Of Apo Betaine Aldehyde Dehydrogenase (Betb) G234S Mutant From Staphylococcus Aureus (Idp00699) With Bme-Modified Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-05-07 Classification: OXIDOREDUCTASE |
 |
2.30 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus With Bme-Free Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-12-11 Classification: OXIDOREDUCTASE |
 |
1.90 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus In Complex With Nad+ And Bme-Free Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-06 Classification: OXIDOREDUCTASE Ligands: NAD, K |
 |
1.85 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus (Idp00699) In Complex With Nad+
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-10-09 Classification: OXIDOREDUCTASE Ligands: NAD, NA |
 |
1.7 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-09-25 Classification: OXIDOREDUCTASE Ligands: MG, CL |
 |
Crystal Structure Of The Uncharacterized Maf Protein Ycef From E. Coli, Mutant D69A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2013-08-14 Classification: HYDROLASE Ligands: UNX |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-03-20 Classification: CELL CYCLE Ligands: GOL, UNX |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2012-10-24 Classification: CELL CYCLE Ligands: UNX |

