Search Count: 25
 |
Crystal Structure Of Exac, An Nad+-Dependent Aldehyde Dehydrogenase, From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, MG |
 |
Crystal Structure Of Exac, An Nad+-Dependent Aldehyde Dehydrogenase, From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-11 Classification: OXIDOREDUCTASE |
 |
Structural Insights Into Bira From Haemophilus Influenzae, A Bifunctional Protein As A Biotin Protein Ligase And A Transcriptional Repressor
Organism: Haemophilus influenzae rd kw20
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: BT5 |
 |
Structural Insights Into Bira From Haemophilus Influenzae, A Bifunctional Protein As A Biotin Protein Ligase And A Transcriptional Repressor
Organism: Haemophilus influenzae rd kw20
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-09-11 Classification: LIGASE Ligands: P33, 1PE |
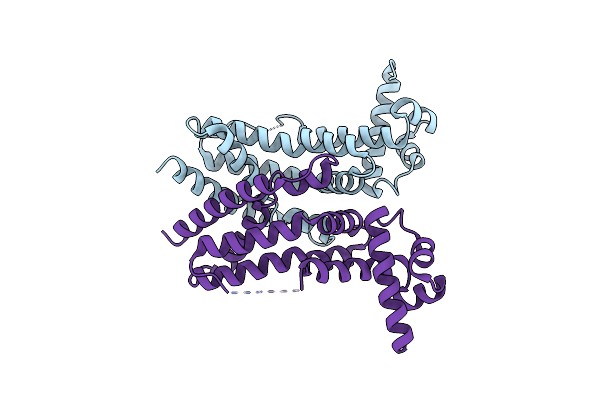 |
Crystal Structure Of Tetr-Type Transcriptional Factor Nalc From P. Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-09-20 Classification: DNA BINDING PROTEIN |
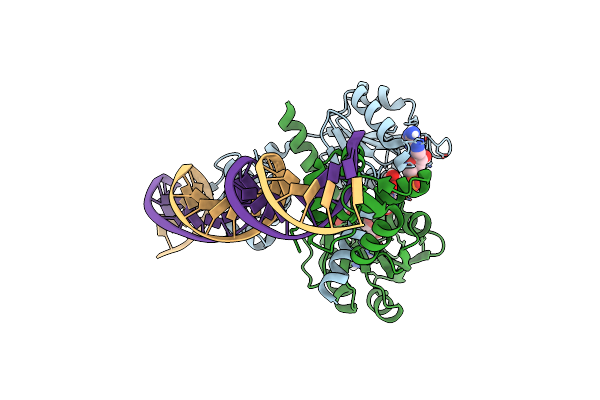 |
Organism: Thermotoga maritima msb8, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-02-16 Classification: GENE REGULATION/DNA Ligands: NAD |
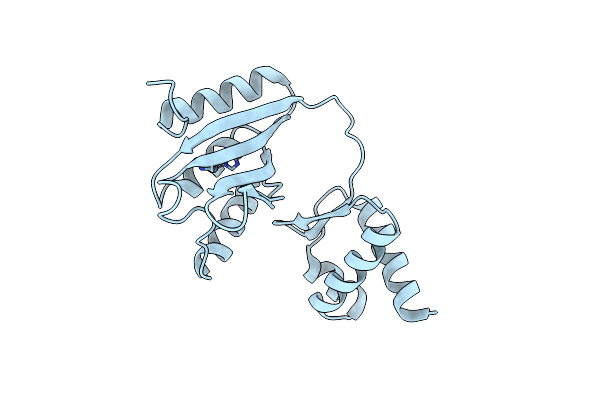 |
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-16 Classification: STRUCTURAL PROTEIN Ligands: ZN |
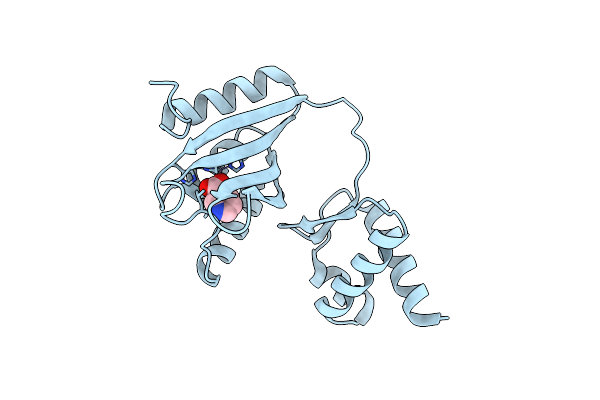 |
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-16 Classification: STRUCTURAL PROTEIN Ligands: NIO, ZN |
 |
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-12-04 Classification: TRANSCRIPTION Ligands: GOL |
 |
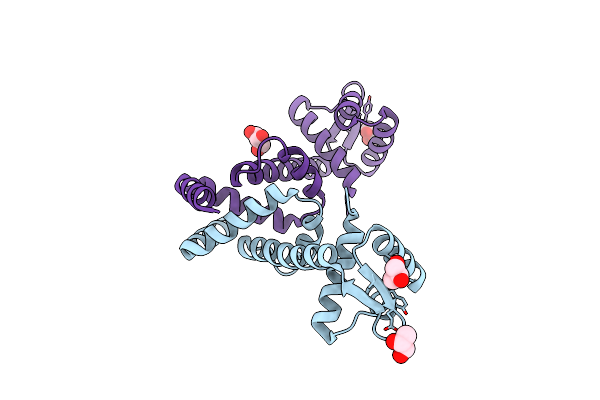
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-12-04 Classification: TRANSCRIPTION Ligands: MG, PO4, MN |
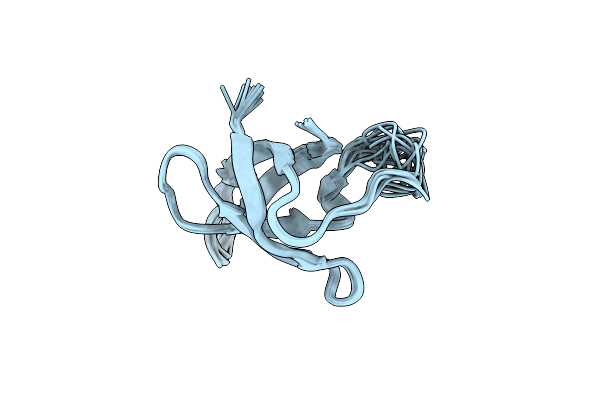 |
Organism: Colwellia psychrerythraea (strain 34h / atcc baa-681)
Method: SOLUTION NMR Release Date: 2018-07-04 Classification: RNA BINDING PROTEIN |
 |
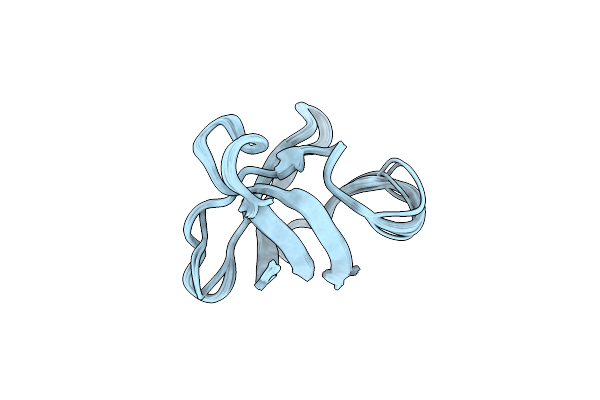
Novel Structural Components Contribute To The High Thermal Stability Of Acyl Carrier Protein From Enterococcus Faecalis
Organism: Enterococcus faecalis v583
Method: SOLUTION NMR Release Date: 2015-12-09 Classification: BIOSYNTHETIC PROTEIN |
 |
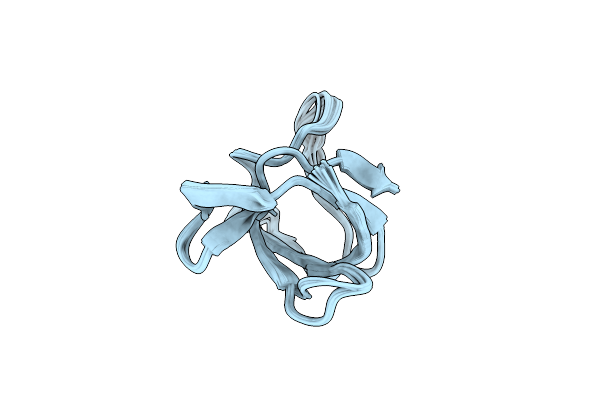
Backbone 1H, 13C, And 15N Chemical Shift Assignments For Cold Shock Protein, Tacsp
Organism: Thermus aquaticus
Method: SOLUTION NMR Release Date: 2014-08-20 Classification: DNA BINDING PROTEIN |
 |
Backbone 1H, 13C, And 15N Chemical Shift Assignments For Cold Shock Protein, Tacsp With Dt7
Organism: Thermus aquaticus
Method: SOLUTION NMR Release Date: 2014-08-20 Classification: DNA BINDING PROTEIN |
 |
 |
Backbone 1H, 13C, And 15N Chemical Shift Assignments For Cold Shock Protein, Lmcsp With Dt7
Organism: Listeria monocytogenes
Method: SOLUTION NMR Release Date: 2013-08-07 Classification: TRANSCRIPTION |
 |
Backbone 1H, 13C, And 15N Chemical Shift Assignments For Cold Shock Protein, Lmcsp
Organism: Listeria monocytogenes
Method: SOLUTION NMR Release Date: 2013-08-07 Classification: TRANSCRIPTION |
 |
Insight Into The Antimicrobial Activities Based On The Structure-Activity Relationships Of Coprisin Isolated From The Dung Beetle, Copris Tripartitus
Organism: Copris tripartitus
Method: SOLUTION NMR Release Date: 2012-11-28 Classification: ANTIMICROBIAL PROTEIN |
 |
Crystal Structure Of Pyrococcus Furiosus Pf2050, A Member Of Duf2666 Family Protein
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2012-04-18 Classification: DNA BINDING PROTEIN Ligands: MPD |
 |
Helical Repeat Structure Of Apoptosis Inhibitor 5 Reveals Protein-Protein Interaction Modules
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-02-22 Classification: APOPTOSIS INHIBITOR |

