Search Count: 63
 |
Organism: Desulfitobacterium hafniense y51
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG |
 |
Photosynthetic Lh1-Rc Complex From The Purple Sulfur Bacterium Allochromatium Vinosum Purified By Sucrose Density
Organism: Allochromatium vinosum dsm 180
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: PHOTOSYNTHESIS Ligands: HEM, MG, Z41, PLM, PGV, BCL, BPH, UQ8, LMT, CDL, FE, MQ8, CRT, CA, LDA |
 |
Photosynthetic Lh1-Rc Complex From The Purple Sulfur Bacterium Allochromatium Vinosum Purified By Ca2+-Deae
Organism: Allochromatium vinosum dsm 180
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: PHOTOSYNTHESIS Ligands: HEM, MG, CA, Z41, PLM, PGV, BCL, BPH, UQ8, FE, MQ8, CRT, CDL, LDA, LMT |
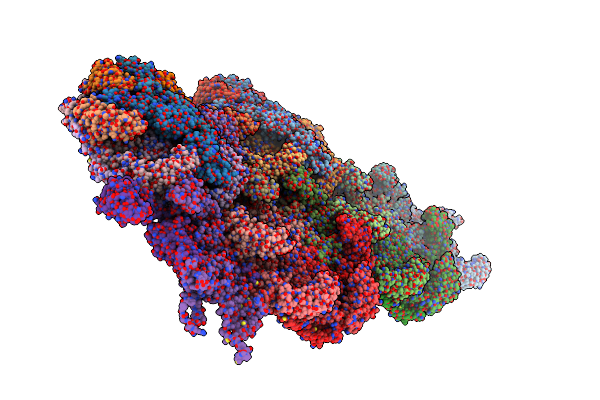 |
Organism: Staphylococcus phage s6
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: VIRUS |
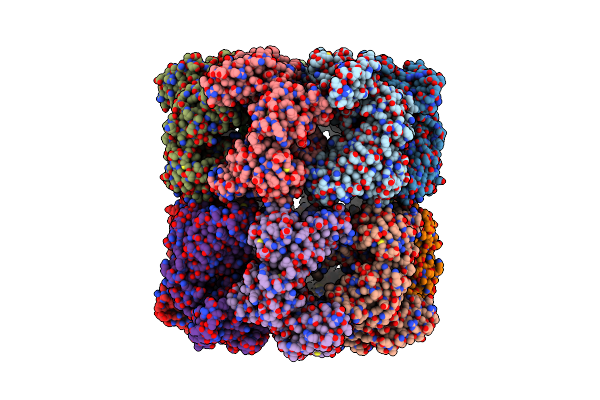 |
Cryo-Em Structure Of Groel Bound To Unfolded Substrate (Ugt1A) At 2.8 Ang. Resolution (Consensus Refinement)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
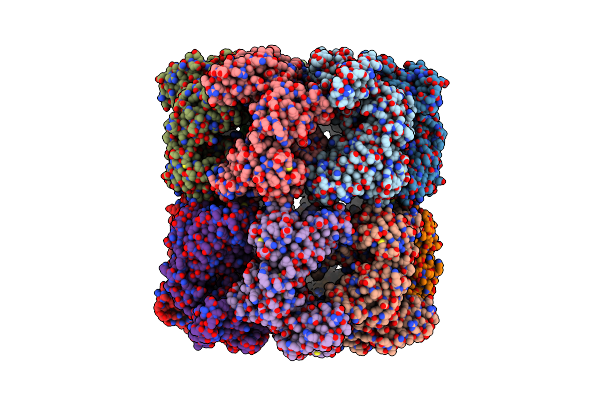 |
Cryo-Em Structure Of Double Occupied Ring (Dor) Of Groel-Ugt1A Complex At 2.7 Ang. Resolution
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
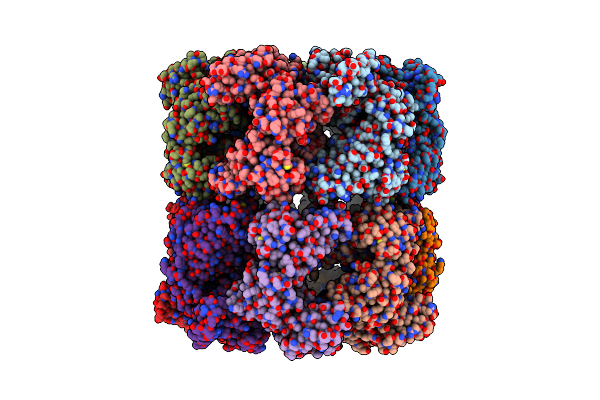 |
Cryo-Em Structure Of Single Empty Ring 2 (Ser2) Of Groel-Ugt1A Complex At 3.2 Ang. Resolution
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 4 (Or4) Of Groel Complexed With Polyalanine Model Of Ugt1A From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Empty Ring Subunit 1 (Er1) From Single Empty Ring Of Groel-Ugt1A Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Empty Ring Subunit 2 (Er2) From Groel-Ugt1A Single Empty Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 1 (Or1) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 2 (Or2) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 3 (Or3) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 4 (Or4) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Organism: Clostridium botulinum bf
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: CELL CYCLE Ligands: ADP, MG |
 |
Organism: Clostridium botulinum
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: CELL CYCLE Ligands: GTP |
 |
Organism: Clostridium botulinum bf
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: CELL CYCLE Ligands: GDP, MG |
 |
Organism: Clostridium botulinum
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: CELL CYCLE Ligands: GTP |
 |
Organism: Helicobacter phage khp30
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: ZN, BAT |

