Search Count: 88
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1991
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: A1ECY, FMN, SF4, MG |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1872
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1EEQ |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: METAL BINDING PROTEIN Ligands: SO4, PGE |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Zea Mays 3-Phosphoglycerate Dehydrogenase S282L Mutant
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Incompetent State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Intermediate State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Nts-Cleaving State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG, GS, SC, AS |
 |
Cryo-Em Structure Of Caslambda2-Crrna-Target Dna Ternary Complex In The Ts-Cleaving State
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Ailuropoda melanoleuca
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSPORT PROTEIN Ligands: NAG, A1L0I |
 |
Organism: Ailuropoda melanoleuca
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSPORT PROTEIN Ligands: NAG, A1L0P |
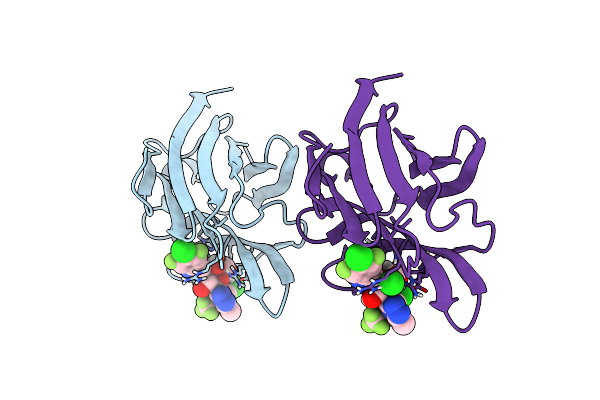 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: A1L0Q, CL |
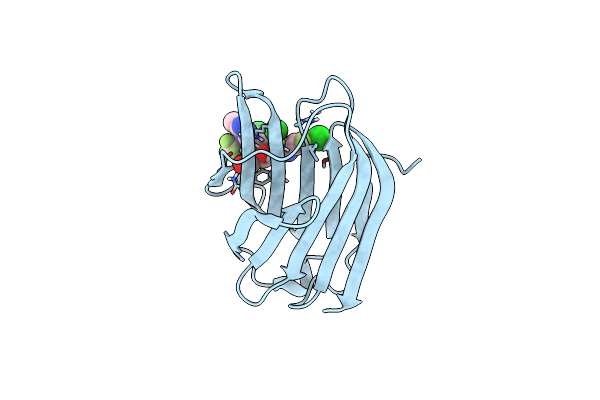 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: A1L0Q, CL |
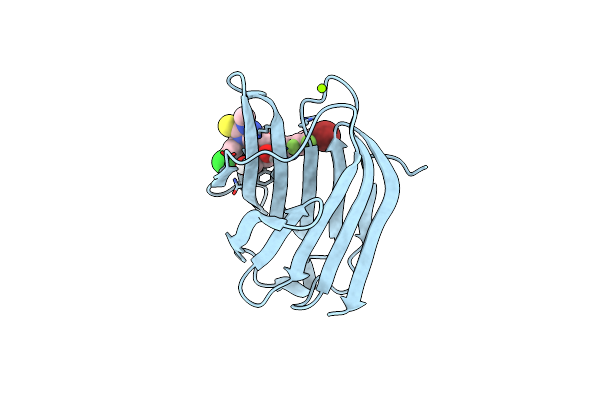 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: MG, A1L0R |
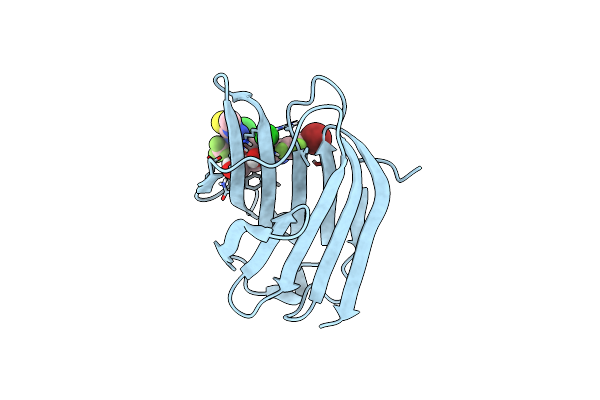 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: A1L0R, CL |
 |
Organism: Erythrobacter cryptus dsm 12079
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: MES |
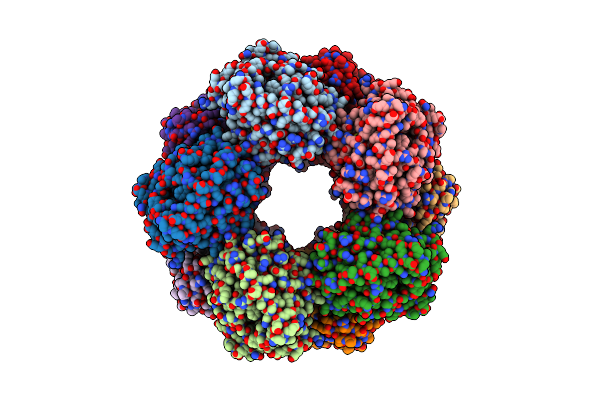 |
Cryoem Structure Of A 2,3-Hydroxycinnamic Acid 1,2-Dioxygenase Mhpb In Substrate Bound Form
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: BIOSYNTHETIC PROTEIN Ligands: FE2, A1EG2 |
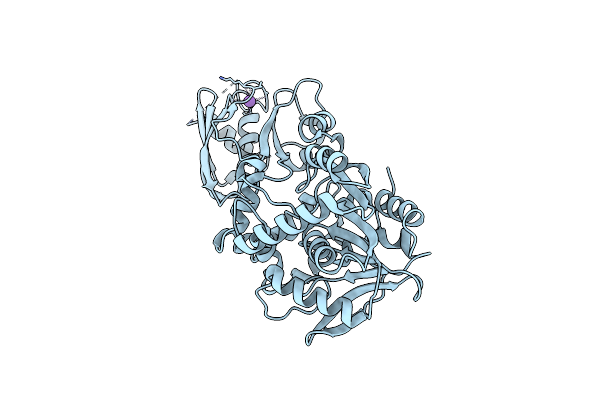 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-12-18 Classification: FLUORESCENT PROTEIN Ligands: K |
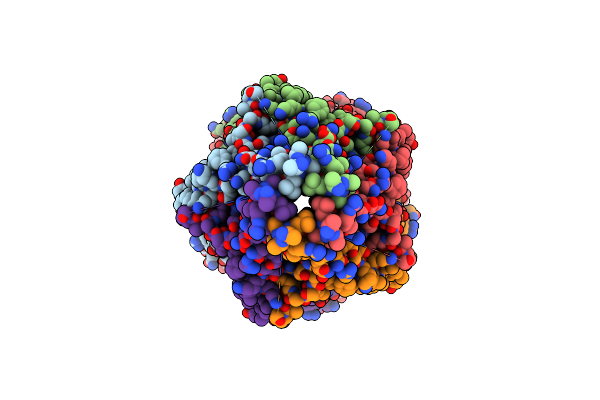 |
Organism: Oryzias latipes
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: TRANSPORT PROTEIN Ligands: NAG |

