Search Count: 206
 |
Crystal Structure Of The Histidine Kinase Vc2136 From Vibrio Cholerae Serotype O1
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: SO4, CL |
 |
Crystal Structure Of C4-Dicarboxylate-Binding Protein (Pa0884) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With L-Malate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: LMR |
 |
Crystal Structure Of C4-Dicarboxylate-Binding Protein (Pa0884) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With Mesaconic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: MEZ, MG, CL |
 |
Crystal Structure Of C4-Dicarboxylate-Binding Periplasmic Protein (Pa5167) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With L-Malate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: LMR, ZN |
 |
Crystal Structure Of Acyl-Coa Lyase Subunit Beta From Pseudomonas Aeruginosa Pao1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE |
 |
Crystal Structure Of The Sars-Cov-2 2'-O-Methyltransferase With (M7Gpppa)Pupu (Cap-0) And S-Adenosyl-L-Homocysteine (Sah).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: TRANSFERASE, Viral Protein Ligands: SAH, CL, SO4, ZN, MGT |
 |
Crystal Structure Of The C-Terminal Cytoplasmic Domain Of Nsp4 From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: VIRAL PROTEIN |
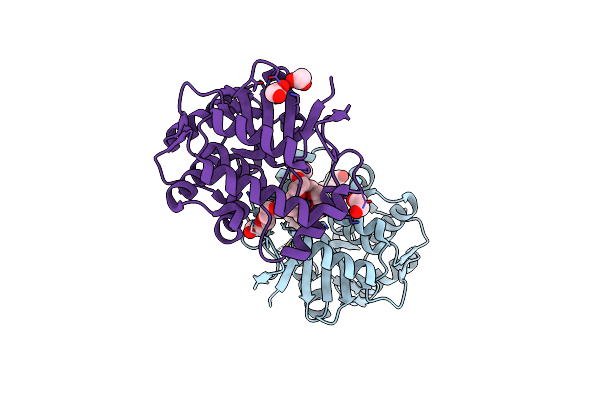 |
Crystal Structure Of The Surface Protein (Cd630_07380) From Clostridium Difficile Strain 630
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN |
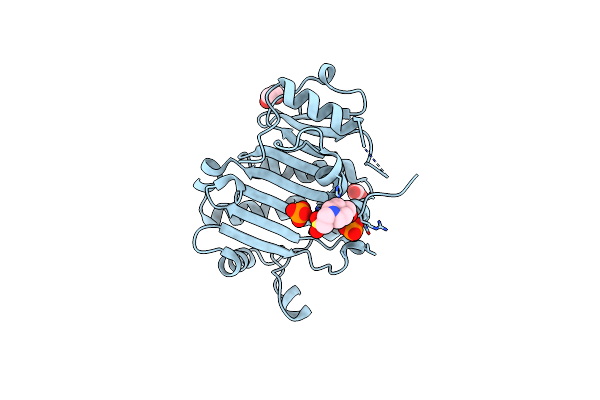 |
Crystal Structure Of The Immunity Protein (52 Domain-Containing Protein) From Pseudomonas Aeruginosa Pao1.
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: ANTITOXIN Ligands: EPE, PO4, EDO, CO3 |
 |
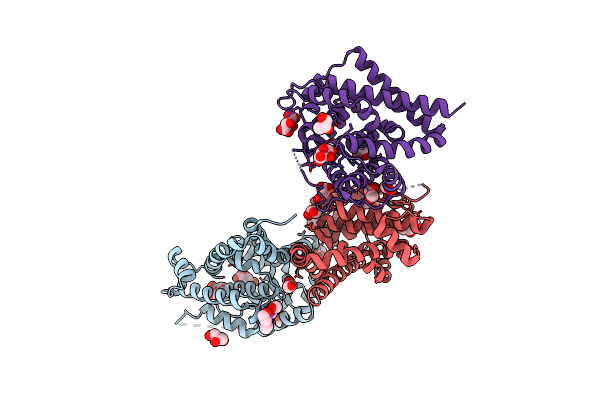
Crystal Structure Of Sh3-Like_Bac-Type Domain (79-145) Of Conserved Domain Protein Gbaa_2967 From Bacillus Anthracis Ames Ancestor
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: UNKNOWN FUNCTION Ligands: MG, FMT |
 |
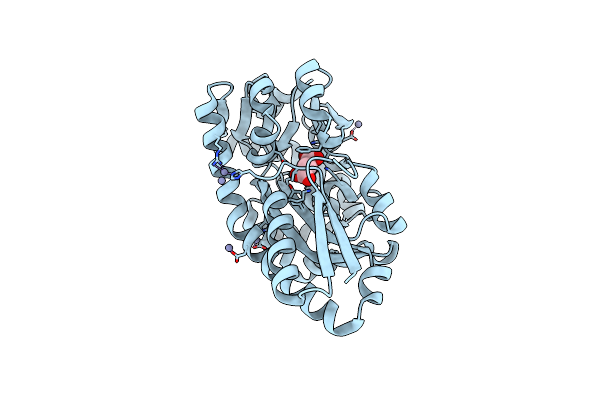
Crystal Structure Of N-Terminal Domain Of Fic Family Protein From Bordetella Bronchiseptica
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2024-10-30 Classification: TRANSFERASE |
 |
Crystal Structure Of C4-Dicarboxylate-Binding Periplasmic Protein (Pa5167) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With Succinic Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-10-09 Classification: TRANSPORT PROTEIN Ligands: SIN, ZN |
 |
Crystal Structure Of C4-Dicarboxylate-Binding Protein (Pa0884) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With Succinic Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-10-09 Classification: TRANSPORT PROTEIN Ligands: SIN, GOL |
 |
High Resolution Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: FMT, SO4 |
 |
Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50 In A Complex With Avibactam
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: NXL, SIN, FMT, EDO |
 |
Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50 In A Complex With Clavulonate
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MLA, TEM, CL |
 |
Crystal Structure Of The Open Unbound Catalytically Inactive Makes Caterpillars Floppy-Like (Mcf) Effector From Vibrio Vulnificus Cmcp6
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: TOXIN Ligands: SO4, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: FE, PO4 |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION Ligands: GOL, EDO |
 |
Crystal Structure Of Nlpc/P60 Domain From Clostridium Innocuum Nlpc/P60 Domain-Containing Protein Ci_01448.
Organism: [clostridium] innocuum 2959
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-10-18 Classification: HYDROLASE Ligands: SO4, EDO, GOL |

