Search Count: 27
 |
Organism: Saccharomyces cerevisiae cen.pk113-7d, Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: DNA BINDING PROTEIN Ligands: ATP, MG |
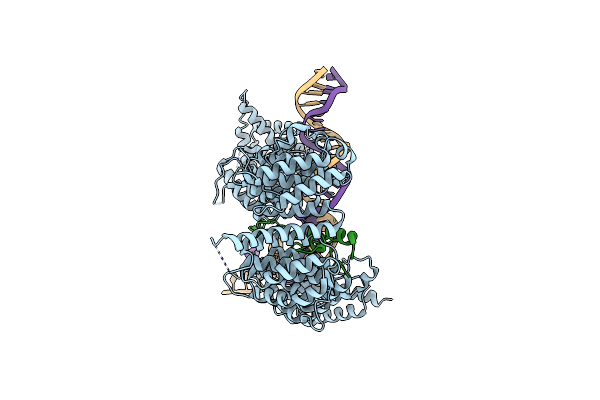 |
Organism: Saccharomyces cerevisiae s288c, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: DNA BINDING PROTEIN |
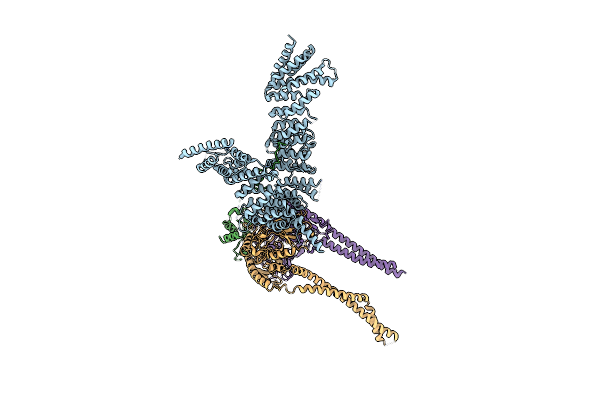 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: CELL CYCLE |
 |
Organism: Chaetomium thermophilum
Method: SOLUTION NMR Release Date: 2019-07-03 Classification: STRUCTURAL PROTEIN |
 |
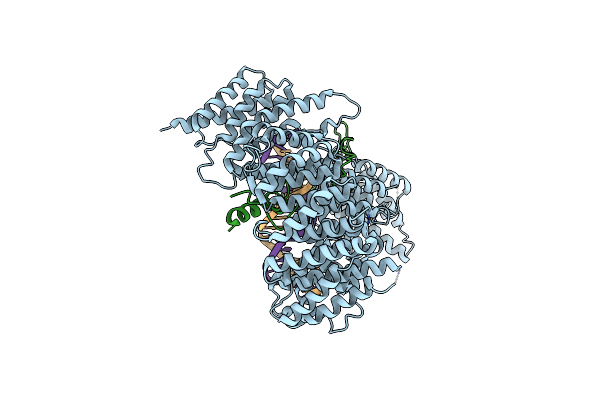
Crystal Structure Of The C. Thermophilum Condensin Smc2 Atpase Head (Crystal Form Ii)
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-03 Classification: CELL CYCLE |
 |
Crystal Structure Of The C. Thermophilum Condensin Smc2 Atpase Head (Crystal From I)
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2019-07-03 Classification: CELL CYCLE |
 |
Crystal Structure Of The C. Thermophilum Condensin Smc4 Atpase Head In Complex With The C-Terminal Domain Of Brn1
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-07-03 Classification: CELL CYCLE Ligands: SO4 |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-07-03 Classification: CELL CYCLE |
 |
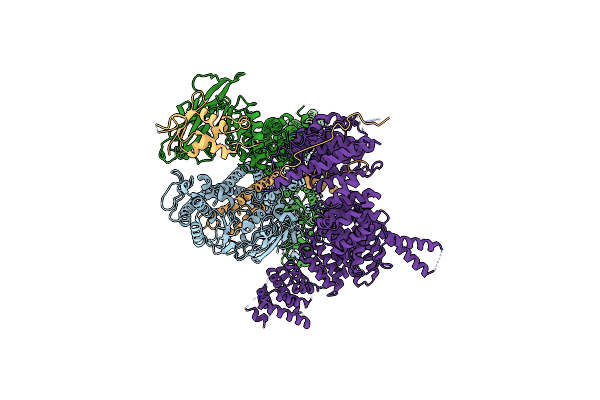
Crystal Structure Of The C. Thermophilum Condensin Ycs4-Brn1 Subcomplex Bound To The Smc4 Atpase Head In Complex With The C-Terminal Domain Of Brn1
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:5.80 Å Release Date: 2019-07-03 Classification: CELL CYCLE |
 |
Crystal Structure Of The S. Cerevisiae Condensin Ycg1-Brn1 Subcomplex Bound To Dna (Short Kleisin Loop)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2017-10-18 Classification: CELL CYCLE |
 |
Crystal Structure Of The S. Cerevisiae Condensin Ycg1-Brn1 Subcomplex Bound To Dna (Crystal Form Ii)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2017-10-18 Classification: CELL CYCLE Ligands: TRS, PGE |
 |
Crystal Structure Of The S. Cerevisiae Condensin Ycg1-Brn1 Subcomplex Bound To Dna (Crystal Form I)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2017-10-18 Classification: CELL CYCLE Ligands: 1PE, SO4 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-10-18 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2017-10-18 Classification: CELL CYCLE |
 |
Structural Basis Of Histone H2A-H2B Recognition By The Essential Chaperone Fact
Organism: Chaetomium thermophilum var. thermophilum, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-05-29 Classification: CHAPERONE/NUCLEAR PROTEIN Ligands: CL, TRS |
 |
Organism: Chaetomium thermophilum var. thermophilum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-05-29 Classification: TRANSCRIPTION/REPLICATION Ligands: SO4 |
 |
Organism: Chaetomium thermophilum var. thermophilum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-29 Classification: TRANSCRIPTION/REPLICATION Ligands: CA, ACT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-04-03 Classification: TRANSCRIPTION |

