Search Count: 15
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The Allosteric Antagonist A438079
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: KE3, GDP, ZN, NAG, PLM, NA |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The Allosteric Antagonist A839977
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: KDU, GDP, ZN, NAG, PLM, NA |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The Allosteric Antagonist Azd9056
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: KEI, GDP, ZN, NAG, PLM, NA |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The Allosteric Antagonist Gsk1482160
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: KF0, GDP, ZN, NAG, PLM, NA |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The Allosteric Antagonist Jnj47965567
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: 7RV, GDP, ZN, NAG, PLM, NA |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The Allosteric Antagonist Methyl Blue
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN Ligands: KFM, GDP, ZN, NAG, PLM |
 |
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: GDP, ZN, NAG, PLM, NA |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In Complex With The High-Affinity Agonist Bzatp
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: KD9, GDP, ZN, NAG, PLM |
 |
Cryo-Em Structure Of The Rat P2X7 Receptor In The Apo Closed State Purified In The Absence Of Sodium
Organism: Rattus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: GDP, ZN, NAG, PLM |
 |
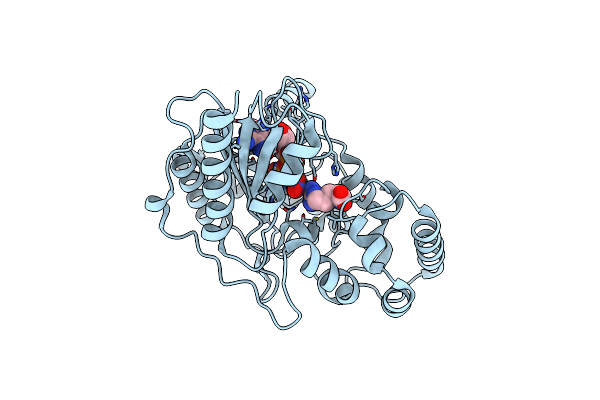
Ambient Temperature Transition State Structure Of Arginine Kinase - Crystal 8/Form I
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: ADP, ARG, NO3, MG |
 |
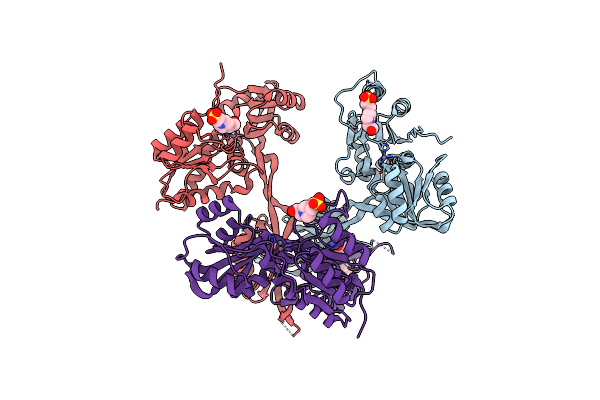
Ambient Temperature Transition State Structure Of Arginine Kinase - Crystal 11/Form Ii
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: NO3, MG, ARG, ADP |
 |
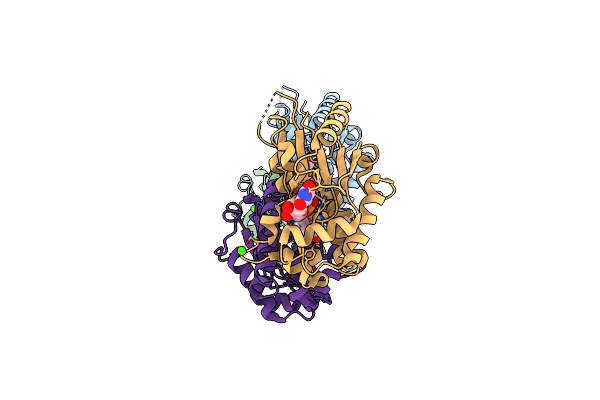
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-05-16 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN, EPE, TRS |
 |
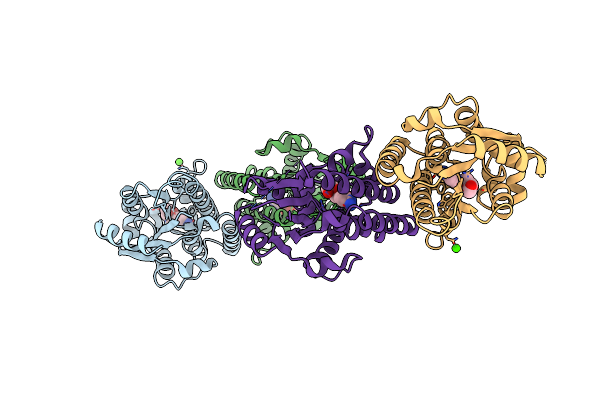
Crystal Structure Of Human Deoxycytidine Kinase In Complex With Deoxycytidine And Uridine Diphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: CA, MG, DCZ, UDP |
 |
Crystal Structure Of Human Deoxycytidine Kinase In Complex With Deoxycytidine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: CA, DCZ |
 |
Crystal Structure Of Mtan, The Bacillus Subtilis Multidrug Transporter Activator, N-Terminus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2001-11-28 Classification: TRANSCRIPTION |

