Search Count: 18
 |
Crystal Structure Of The Essential Dimeric Lysa From Phaeodactylum Tricornutum
Organism: Phaeodactylum tricornutum
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2021-06-16 Classification: LYASE Ligands: DLY, SO4 |
 |
Crystal Structure Of A Marine Metagenome Trap Solute Binding Protein Specific For Pyroglutamate (Sorcerer Ii Global Ocean Sampling Expedition, Unidentified Microbe, Scf7180008839099) In Complex With Co-Purified Pyroglutamate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-04-29 Classification: SUGAR BINDING PROTEIN Ligands: PCA, EDO, CL, FMT |
 |
Crystal Structure Of A Marine Metagenome Trap Solute Binding Protein Specific For Aromatic Acid Ligands (Sorcerer Ii Global Ocean Sampling Expedition, Unidentified Microbe, Locus Tag Gos_1523157) In Complex With Co-Purified 4-Hydroxybenzoate
Organism: Uncultured marine bacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-01-18 Classification: TRANSPORT PROTEIN Ligands: PHB, PEG, BR |
 |
Crystal Structure Of A Marine Metagenome Trap Solute Binding Protein Specific For Aromatic Acid Ligands (Sorcerer Ii Global Ocean Sampling Expedition, Unidentified Microbe, Locus Tag Gos_1523157) In Complex With Co-Crystallized 3-Hydroxybenzoate
Organism: Uncultured marine bacterium
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-01-18 Classification: TRANSPORT PROTEIN Ligands: 3HB, PEG |
 |
Crystal Structure Of A Marine Metagenome Trap Solute Binding Protein Specific For Aromatic Acid Ligands (Sorcerer Ii Global Ocean Sampling Expedition, Unidentified Microbe, Locus Tag Gos_1523157, Triple Surface Mutant K158A_K223A_K313A) In Complex With Co-Purified Parahydroxybenzoate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-01-18 Classification: TRANSPORT PROTEIN Ligands: PEG, TRS, PHB |
 |
Crystal Structure Of A Marine Metagenome Trap Solute Binding Protein Specific For Aromatic Acid Ligands (Sorcerer Ii Global Ocean Sampling Expedition, Unidentified Microbe, Locus Tag Gos_1523157, Triple Surface Mutant K158A_K223A_K313A) In Complex With Metahydroxyphenylacetate, Thermal Exchange Of Ligand
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-01-18 Classification: TRANSPORT PROTEIN Ligands: 3HP, PEG |
 |
The Structure Of A Family 4 Acetyl Xylan Esterase From Clostridium Thermocellum In Complex With A Magnesium Ion.
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2006-01-23 Classification: HYDROLASE Ligands: MG |
 |
The Structure Of A Family 4 Acetyl Xylan Esterase From Clostridium Thermocellum In Complex With A Colbalt Ion.
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-01-23 Classification: HYDROLASE Ligands: CO |
 |
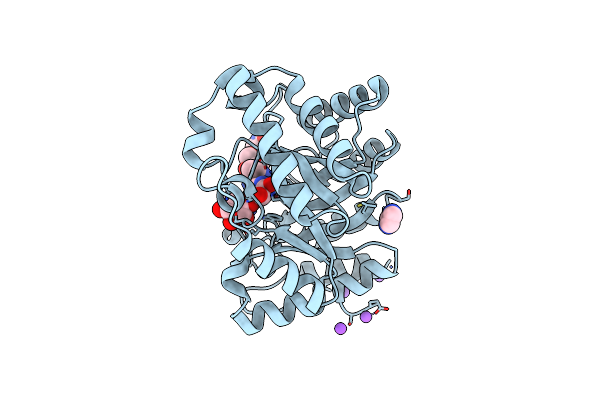
Family 4 Carbohydrate Esterase From Streptomyces Lividans In Complex With Acetate
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-01-23 Classification: HYDROLASE Ligands: ZN, ACT |
 |
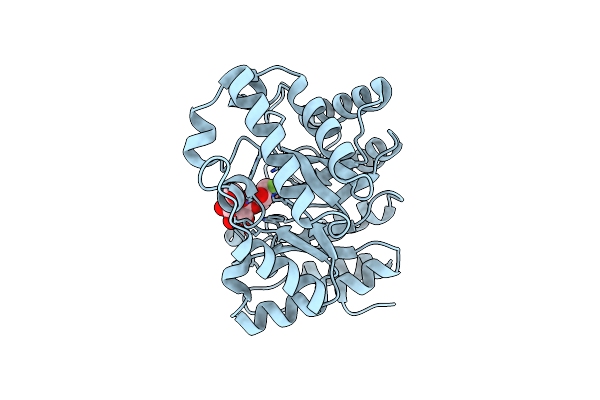
Xylanase Xyn10A From Streptomyces Lividans In Complex With Xylobio-Isofagomine Lactam
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2003-04-08 Classification: HYDROLASE/LECTIN |
 |
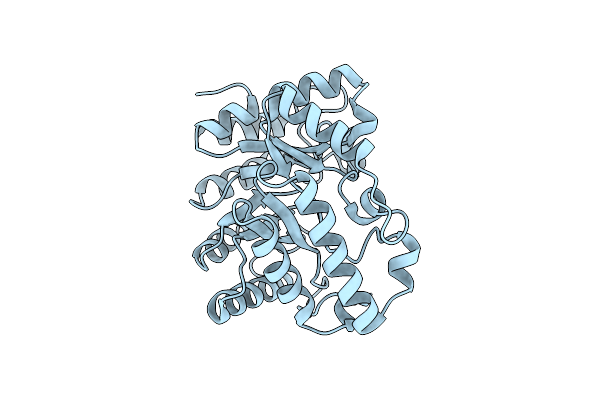
Xylanase 10A From Sreptomyces Lividans. Cellobiosyl-Enzyme Intermediate At 1.7 A
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2001-04-05 Classification: HYDROLASE |
 |
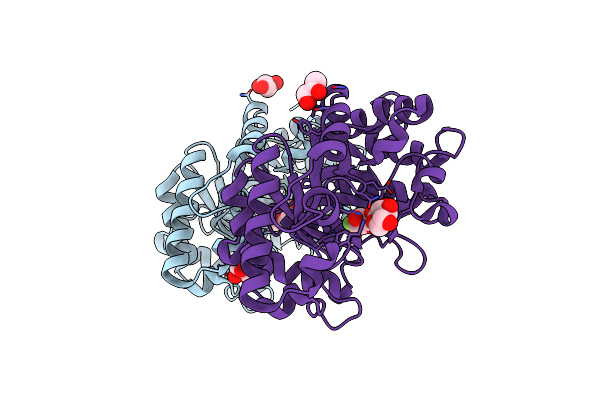
Xylanase 10A From Sreptomyces Lividans. Native Structure At 1.2 Angstrom Resolution
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2001-04-05 Classification: HYDROLASE |
 |
Xylanase 10A From Sreptomyces Lividans. Xylobiosyl-Enzyme Intermediate At 1.65 A
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2001-04-05 Classification: HYDROLASE Ligands: GOL |
 |
Catalysis And Specificity In Enzymatic Glycoside Hydrolases: A 2,5B Conformation For The Glycosyl-Enzyme Intermidiate Revealed By The Structure Of The Bacillus Agaradhaerens Family 11 Xylanase
Organism: Bacillus agaradhaerens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-05-17 Classification: HYDROLASE |
 |
Catalysis And Specificity In Enzymatic Glycoside Hydrolases: A 2,5B Conformation For The Glycosyl-Enzyme Intermidiate Revealed By The Structure Of The Bacillus Agaradhaerens Family 11 Xylanase
Organism: Bacillus agaradhaerens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2000-05-17 Classification: HYDROLASE Ligands: XYP |
 |
 |
Streptomyces Lividans Endoglucanase (Ec: 3.2.1.4) Complex With Modified Glucose Trimer
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 1999-11-10 Classification: HYDROLASE |
 |
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 1998-11-25 Classification: ENDOGLUCANASE |

