Search Count: 20
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: CL |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: PGO |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: MPD |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State The With Methylated Lysines
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
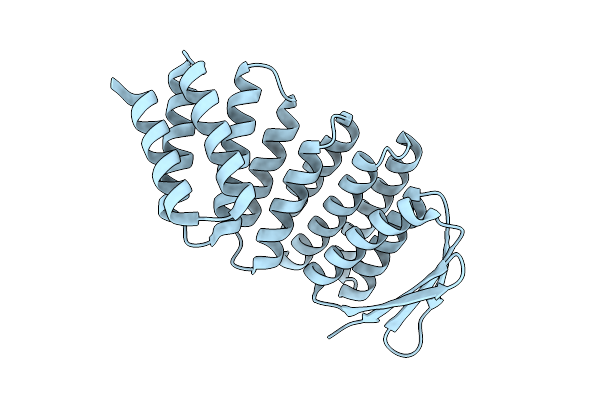 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
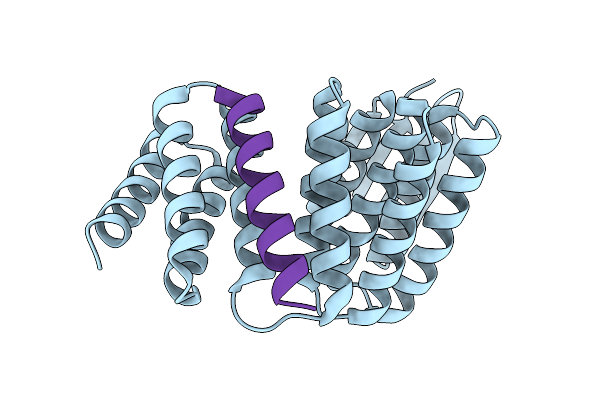 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Facilitated Dissociation Target Lhd101An1: Lhd101A With An N-Terminal Extension
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
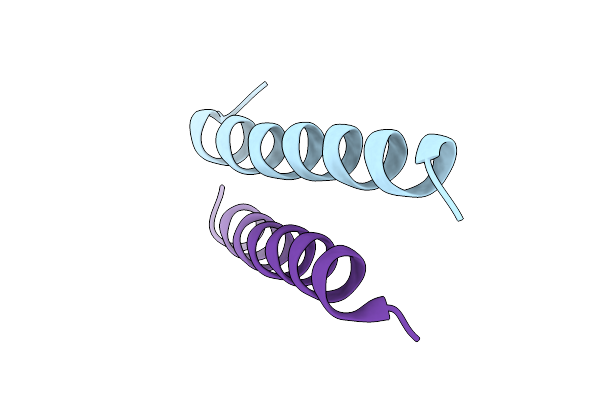 |
Crystal Structure Of Designed Conformational Switch Effector Peptide Cs221B
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
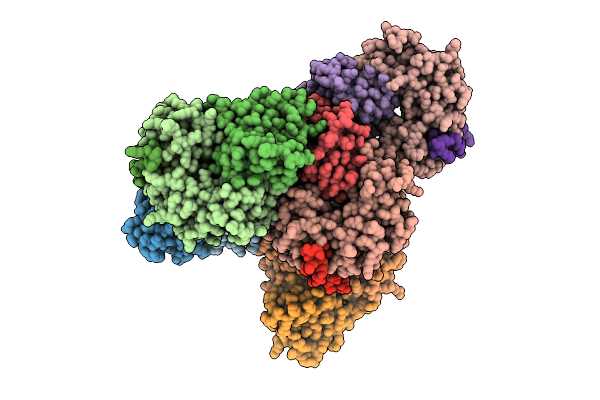 |
Designed Allosteric Facilitated Dissociation Switch As1_K46L_E50W_K172W_E173Y In Complex State The
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: ONCOPROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-09-25 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: DE NOVO PROTEIN Ligands: HEM |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
Structure Of Ytm1 Bound To The C-Terminal Domain Of Erb1 In P21 21 2 Space Group
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-11-04 Classification: PROTEIN BINDING Ligands: EDO, NA, GOL, CL |
 |
Structure Of Ytm1 Bound To The C-Terminal Domain Of Erb1 In P 65 2 2 Space Group
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-10-28 Classification: PROTEIN BINDING Ligands: CL |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-10-28 Classification: PROTEIN BINDING Ligands: CL |

