Search Count: 17
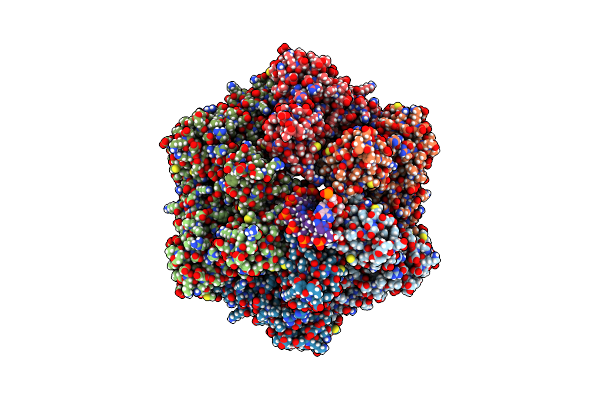 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
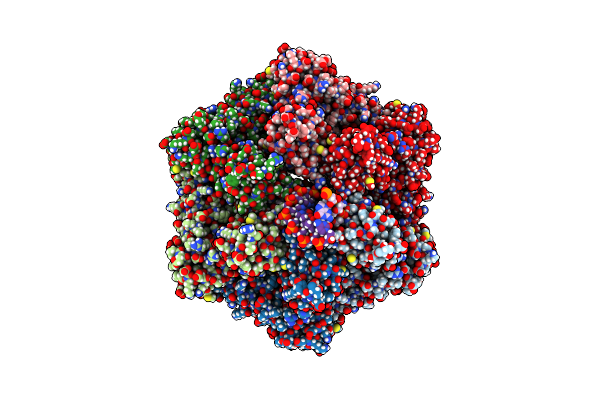 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S2 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
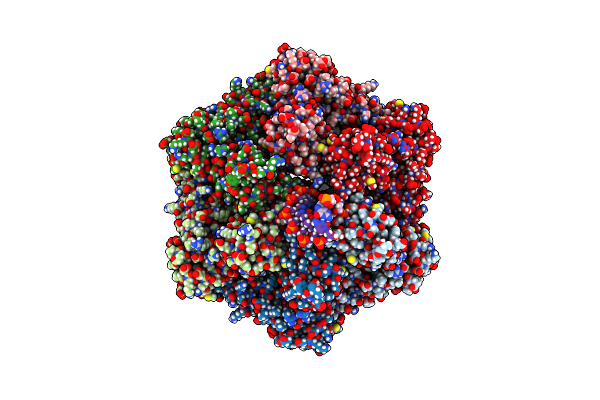 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, MG, AGS |
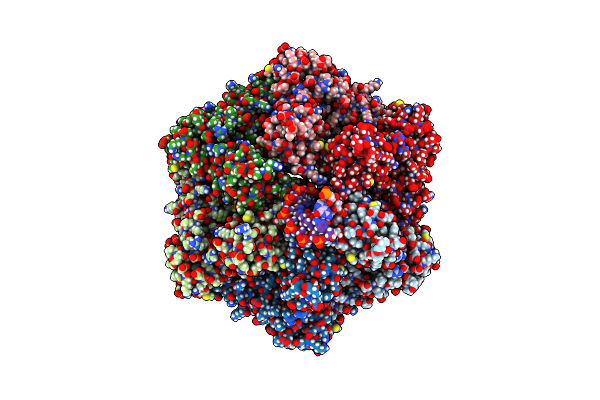 |
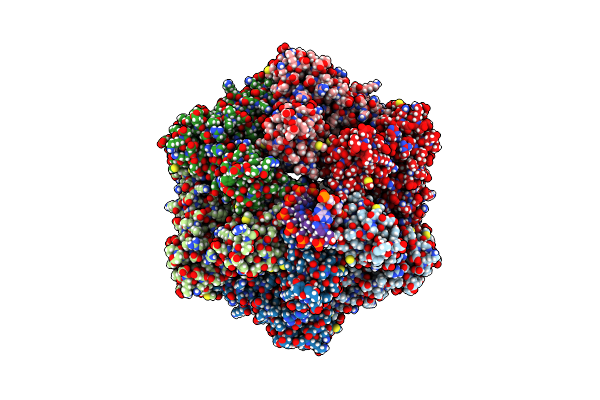
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S4 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
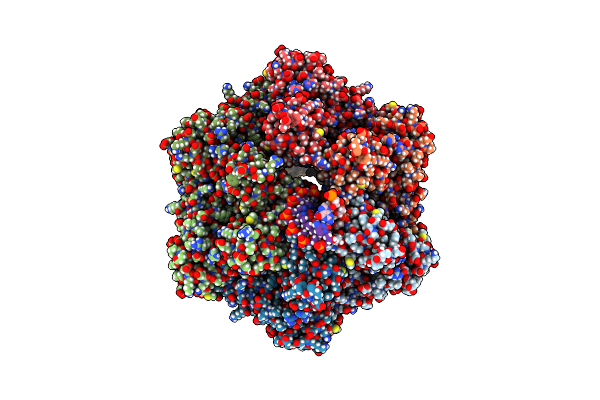
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S5 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
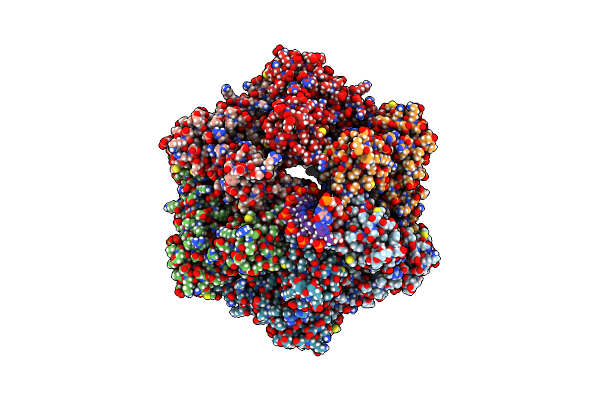
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
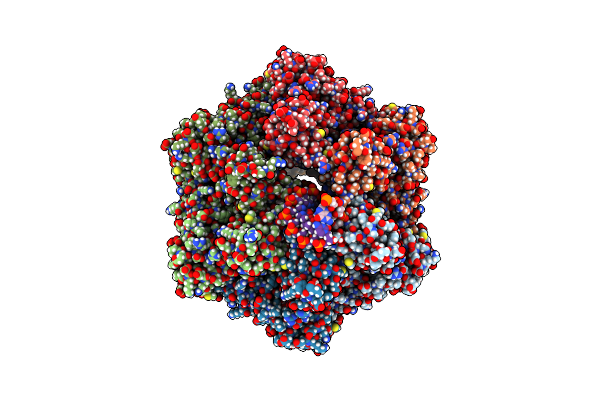
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0-A [T2 Dataset]
Organism: Streptococcus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruva-Hj Core [T2 Dataset]
Organism: Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-08-26 Classification: STRUCTURAL PROTEIN Ligands: CA, MG, TRS |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-08-26 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2020-08-26 Classification: STRUCTURAL PROTEIN |
 |
The Crystal Structure Of The Two Spectrin Repeat Domains From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-16 Classification: STRUCTURAL PROTEIN Ligands: BME |
 |
The Crystal Structure Of The Actin Binding Domain (Abd) Of Alpha Actinin From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2018-05-16 Classification: STRUCTURAL PROTEIN Ligands: CA, TRS |
 |
Structure And Conformational Variability Of The Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:20.00 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: FMN |
 |
Structure And Conformational Variability Of The Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:17.50 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: FMN |

