Search Count: 65
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Structure Of Aldo-Keto Reductase 1C3 (Akr1C3) In Complex With An Inhibitor Meds765
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-08-21 Classification: OXIDOREDUCTASE Ligands: NAP, A1ICG, EDO, MES |
 |
Crystal Structure Of Trka D5 Domain In Complex With Two Different Macrocyclic Peptides
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: EDO |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: EDO, GM1 |
 |
Crystal Structure Of Human Dihydroorotate Dehydrogenase In Complex With The Inhibitor 2-Hydroxy-N-(2-Isopropyl-5-Methyl-4-(Pyridin-4-Yloxy)Phenyl)Pyrazolo[1,5-A]Pyridine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-06-19 Classification: FLAVOPROTEIN Ligands: FMN, ORO, YMH, ACT, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: TRANSPORT PROTEIN Ligands: MG, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: TRANSPORT PROTEIN Ligands: PGW, MG, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: TRANSPORT PROTEIN Ligands: MG, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Structure Of Aldo-Keto Reductase 1C3 (Akr1C3) In Complex With An Inhibitor M689, With The 3-Hydroxy-Benzoisoxazole Moiety. Resolution 2.0A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-06 Classification: OXIDOREDUCTASE Ligands: NAP, YMC, EDO |
 |
Crystal Structure Of The Full-Length Dihydroorotate Dehydrogenase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2023-09-06 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSPORT PROTEIN Ligands: MG, CA |
 |
Crystal Structure Of Human Dihydroorotate Dehydrogenase In Complex With The Inhibitor 2-Hydroxy-N-(2-Ispropyl-5-Methyl-4-Phenoxyphenyl)Pyrazolo[1,5-A]Pyridine-3-Carboxamide.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-12 Classification: OXIDOREDUCTASE Ligands: FMN, ORO, IEQ, P6G, ACT |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-02-02 Classification: HYDROLASE |
 |
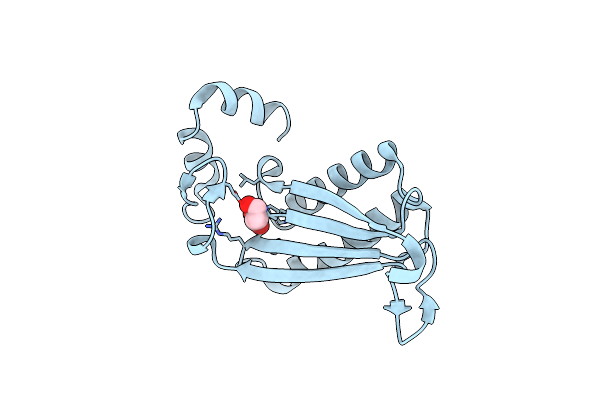
Crystal Structure Of H28A Mutant Of Cg10062 With A Covalent Intermediate Of The Hydration Of Acetylenecarboxylic Acid
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: 3OH |
 |
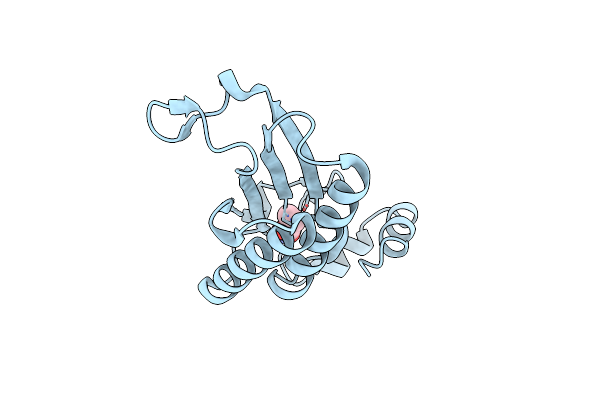
Crystal Structure Of R73A Mutant Of Cg10062 With A Covalent Intermediate Of The Hydration Of Acetylenecarboxylic Acid
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: 3OH |
 |
Crystal Structure Of Y103F Mutant Of Cg10062 With A Covalent Intermediate Of The Hydration Of Acetylenecarboxylic Acid
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-02-02 Classification: ISOMERASE Ligands: 3OH |

