Search Count: 16
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-07-15 Classification: LIPID BINDING PROTEIN Ligands: PGE |
 |
Organism: Filobasidiella neoformans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-05-04 Classification: LYASE Ligands: HG, EMC, SO4, GOL, ACY |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-03-02 Classification: LYASE |
 |
The Crystal Structure Of The First Enzyme Of Pantothenate Biosynthetic Pathway, Ketopantoate Hydroxymethyltransferase From Mycobacterium Tuberculosis Shows A Decameric Assembly And Terminal Helix-Swapping
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-07-15 Classification: TRANSFERASE Ligands: MG |
 |
Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-06-17 Classification: TRANSFERASE, LYASE Ligands: SO4, NI, POP |
 |
Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-06-17 Classification: TRANSFERASE, LYASE Ligands: NI, 1PR |
 |
Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-06-17 Classification: TRANSFERASE, LYASE Ligands: NI, SO4, POP |
 |
Hinge-Bending Motion Of D-Allose Binding Protein From Escherichia Coli: Three Open Conformations
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2003-03-06 Classification: SUGAR BINDING PROTEIN Ligands: NI |
 |
Hinge-Bending Motion Of D-Allose Binding Protein From Escherichia Coli: Three Open Conformations
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2003-03-06 Classification: PERIPLASMIC BINDING PROTEIN Ligands: ZN |
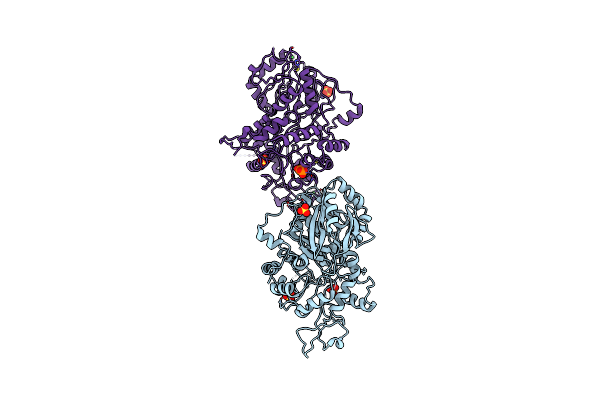 |
Crystal Structure Of Imidazole Glycerol Phosphate Synthase: A Tunnel Through A (Beta/Alpha)8 Barrel Joins Two Active Sites
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2001-10-12 Classification: TRANSFERASE Ligands: NI, SO4, POP |
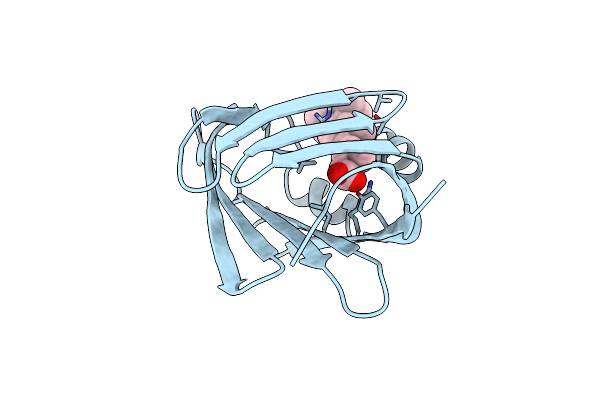 |
Cellular Retinoic Acid Binding Protein Ii In Complex With A Synthetic Retinoic Acid (Ro-13 6307)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1999-12-22 Classification: TRANSPORT PROTEIN Ligands: R13 |
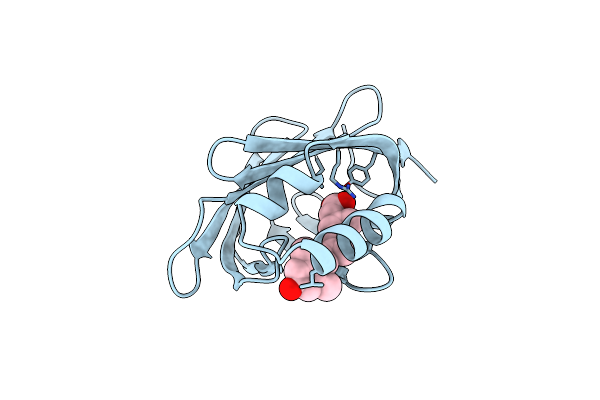 |
Cellular Retinoic Acid Binding Protein Ii In Complex With A Synthetic Retinoic Acid (Ro-12 7310)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-12-22 Classification: TRANSPORT PROTEIN Ligands: R12 |
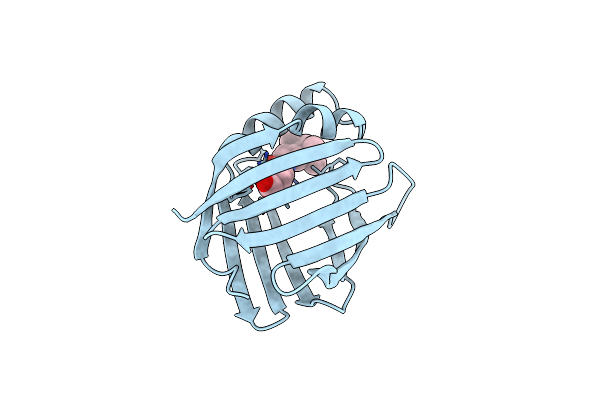 |
Cellular Retinoic Acid Binding Protein I In Complex With A Retinobenzoic Acid (Am80)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1999-12-21 Classification: TRANSPORT PROTEIN Ligands: A80 |
 |
The Crystal Structures Of A2U-Globulin And Its Complex With A Hyaline Droplet Inducer.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1999-08-26 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structures Of A2U-Globulin And Its Complex With A Hyaline Droplet Inducer.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1999-08-13 Classification: LIPID BINDING PROTEIN Ligands: LEO |
 |
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1999-02-16 Classification: PERIPLASMIC SUGAR RECEPTOR Ligands: ZN, SO4, ALL |

