Search Count: 49
 |
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-04-21 Classification: BIOSYNTHETIC PROTEIN Ligands: STE, FAD |
 |
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-04-21 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, PJ8, CO2, STE |
 |
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-04-21 Classification: BIOSYNTHETIC PROTEIN Ligands: STE, FAD |
 |
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-04-21 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, PJ8, CO2, STE, 1PE |
 |
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-04-21 Classification: BIOSYNTHETIC PROTEIN Ligands: STE, FAD, PG4 |
 |
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-04-21 Classification: BIOSYNTHETIC PROTEIN Ligands: STE, FAD |
 |
Dark State Structure Of The C432S Mutant Of Fatty Acid Photodecarboxylase (Fap)
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: STE, FAD, GOL |
 |
Crystal Structure Of Fatty Acid Photodecarboxylase In The Dark State Determined By Serial Femtosecond Crystallography At Room Temperature
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
Structure Of Fatty Acid Photodecarboxylase In Complex With Fad And Palmitic Acid
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, PLM |
 |
Structure Of A Nanobody (Vhh) From Camel Against Phage Tuc2009 Rbp (Bppl, Orf53)
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-12-30 Classification: IMMUNE SYSTEM |
 |
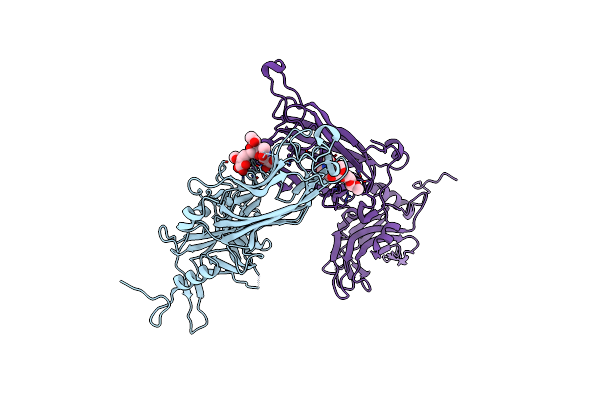
Complex Between Lactococcal Phage Tuc2009 Rbp Head Domain And A Nanobody (L06)
Organism: Camelus dromedarius, Lactococcus phage tuc2009
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-30 Classification: VIRAL PROTEIN |
 |
Structure Of The Tripod (Bppuct-A-L) From The Baseplate Of Bacteriophage Tuc2009
Organism: Lactococcus phage tuc2009
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-12-30 Classification: VIRAL PROTEIN Ligands: SO4, CA |
 |
Phage 1358 Receptor Binding Protein In Complex With The Trisaccharide Glcnac-Galf-Glcome
Organism: Lactococcus phage 1358
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-20 Classification: VIRAL PROTEIN |
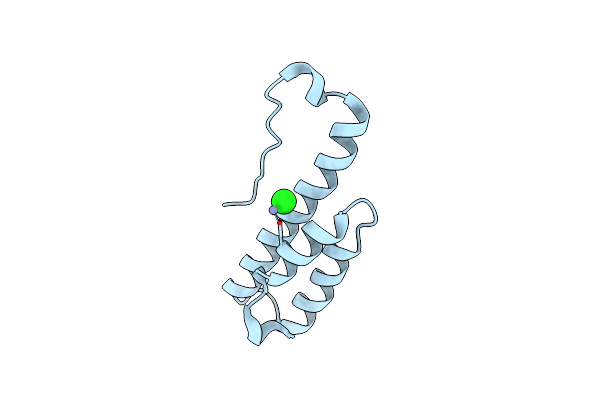 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-03-11 Classification: CONTRACTILE PROTEIN Ligands: ZN, CL |
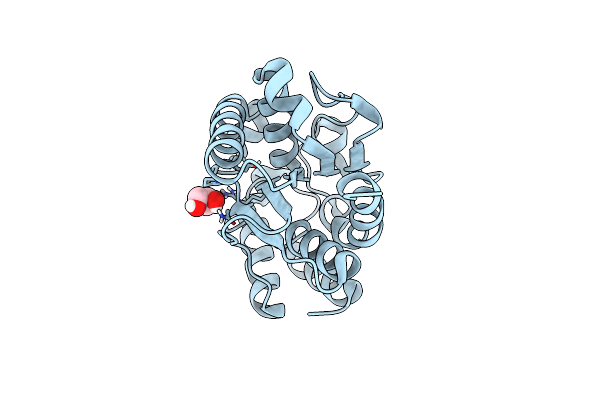 |
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: GOL |
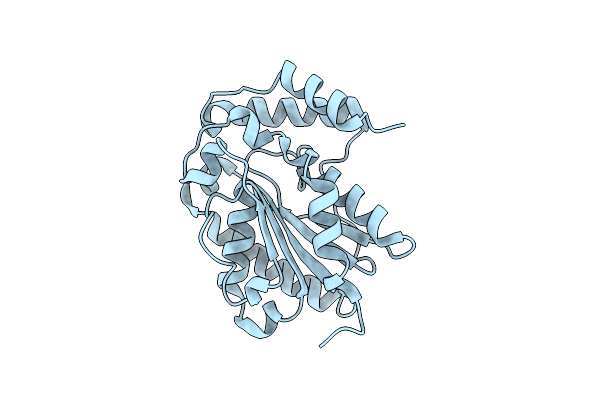 |
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2014-09-17 Classification: HYDROLASE |
 |
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2014-09-17 Classification: hydrolase/hydrolase inhibitor Ligands: C11, BOG |
 |
The Structure Of A 1.8 Mda Viral Genome Injection Device Suggests Alternative Infection Mechanisms
Organism: Lactococcus phage tp901-1
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2014-07-09 Classification: VIRAL PROTEIN |
 |
Structure Of The Rbp From Lactococcal Phage 1358 In Complex With 2 Glcnac Molecules
Organism: Lactococcus phage 1358
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-04-30 Classification: VIRAL PROTEIN Ligands: NDG, NAG, ZN |
 |
Structure Of The Rbp Of Lactococcal Phage 1358 In Complex With Glucose-1-Phosphate
Organism: Lactococcus phage 1358
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2014-04-30 Classification: VIRAL PROTEIN Ligands: G1P |

