Search Count: 7
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: ZN, ZPG |
 |
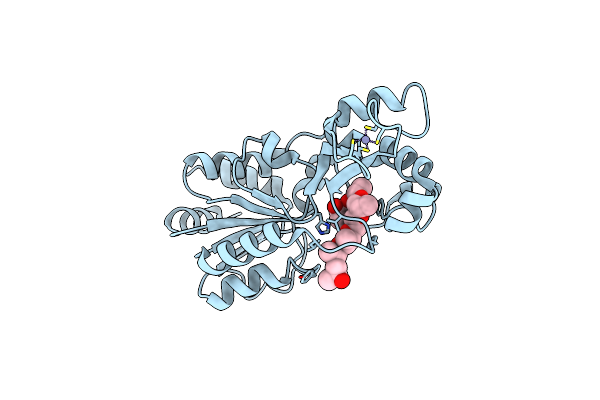
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2006-11-28 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-09-19 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2005-04-26 Classification: HYDROLASE Ligands: ZN, NCA |