Search Count: 40
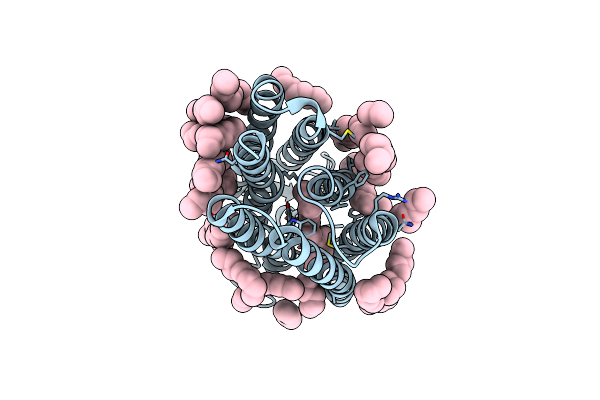 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 20Ms Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, NA |
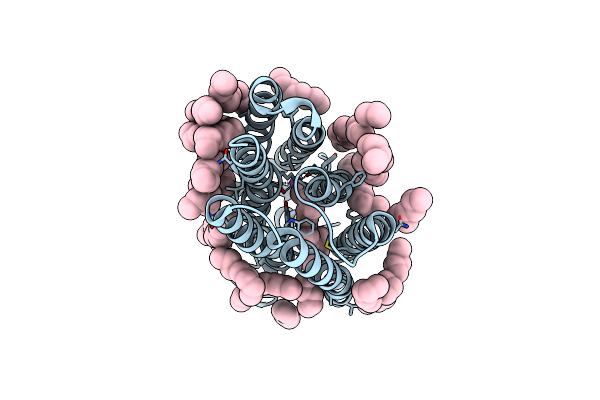 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 1Ms Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, NA |
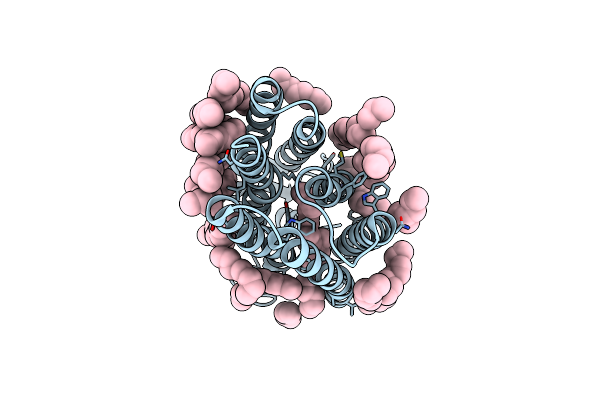 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 30Us+150Us Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
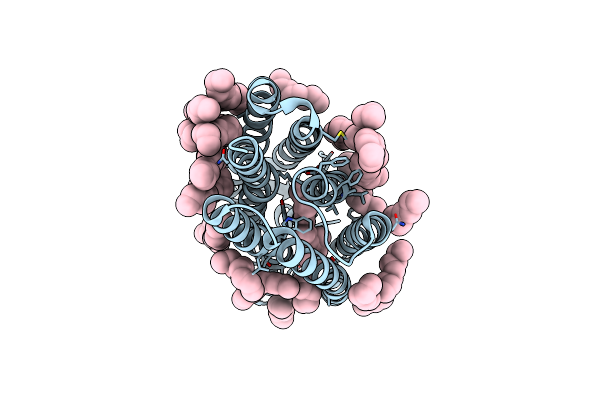 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 1Ns+16Ns Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 800Fs+2Ps Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: Dark Structure In Neutral Conditions With Attached Light Datasets At 800Fs, 2Ps, 100Ps, 1Ns, 16Ns, 1Us, 30Us, 150Us, 1Ms And 20Ms
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: Dark Structure In Acidic Conditions
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Structures Of The Human Ox1 Orexin Receptor Bound To Selective And Dual Antagonists
Organism: Homo sapiens, Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-03-09 Classification: SIGNALING PROTEIN Ligands: SUV, OLA |
 |
Structures Of The Human Ox1 Orexin Receptor Bound To Selective And Dual Antagonists
Organism: Homo sapiens, Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2016-03-09 Classification: SIGNALING PROTEIN Ligands: 4OT, OLA |
 |
Tyr258Phe Mutant Of Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.55A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-10-20 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-06-02 Classification: HYDROLASE Ligands: NAG |
 |
Na+-Dependent Allostery Mediates Coagulation Factor Protease Active Site Selectivity
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2008-03-11 Classification: HYDROLASE Ligands: SO4, BEN, NA, CA |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:0.89 Å Release Date: 2007-12-11 Classification: ELECTRON TRANSPORT Ligands: CU, PO4 |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2007-12-11 Classification: ELECTRON TRANSPORT Ligands: CU1, PO4 |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:0.75 Å Release Date: 2007-08-14 Classification: ELECTRON TRANSPORT Ligands: CU, PO4, NA |
 |
Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Closed Form At 1.15 A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2007-07-31 Classification: OXIDOREDUCTASE |
 |
Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.9A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-07-31 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-07-31 Classification: FLAVOPROTEIN Ligands: FAD, BEZ |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2007-03-13 Classification: ELECTRON TRANSPORT Ligands: CU, PO4 |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2007-03-13 Classification: ELECTRON TRANSPORT Ligands: CU1 |

