Search Count: 7
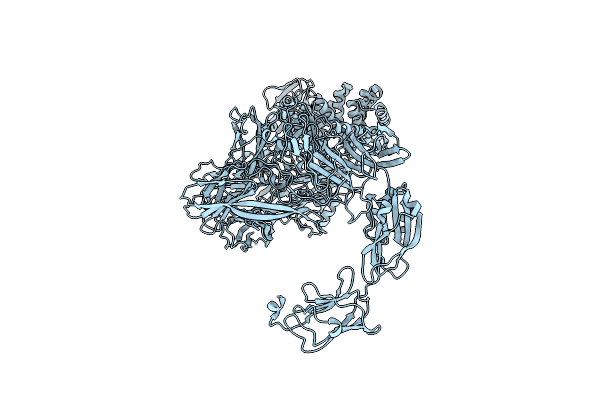 |
Cryo-Em Single Particle 3D Reconstruction Of The Native Conformation Of E. Coli Alpha-2-Macroglobulin (Ecam)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:16.00 Å Release Date: 2015-07-29 Classification: HYDROLASE INHIBITOR |
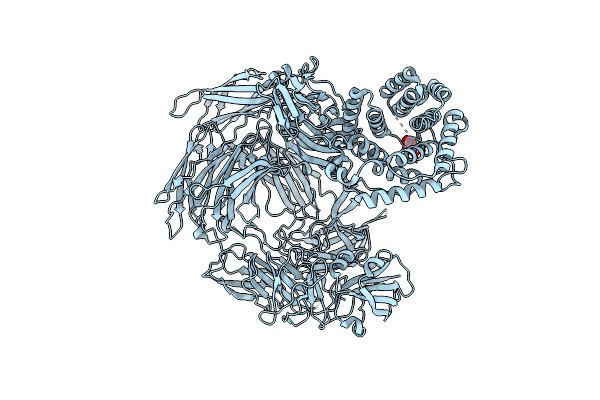 |
Crystal Structure Of Trypsin Activated Alpha-2-Macroglobulin From Escherichia Coli.
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-06-10 Classification: MEMBRANE PROTEIN Ligands: GOL, CL |
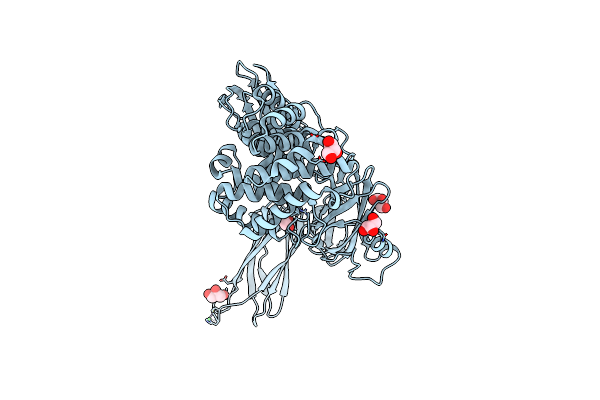 |
Crystal Structure Of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning The Residues From Domain Mg7 To The C-Terminus.
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-06-10 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: NI, GOL |
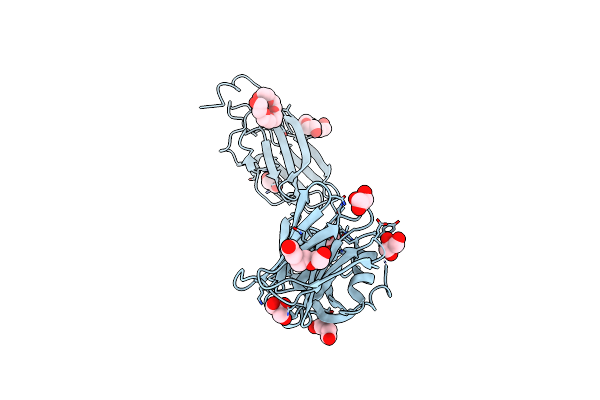 |
Crystal Structure Of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning Domains Mg0-Nie-Mg1.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-06-10 Classification: MEMBRANE PROTEIN Ligands: PG4, PGE, 1PE, PEG, GOL |
 |
Crystal Structure Of Native Alpha-2-Macroglobulin From Escherichia Coli Spanning Domains Nie-Mg1.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-06-10 Classification: MEMBRANE PROTEIN Ligands: ACT, GOL |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-06-12 Classification: PROTEIN BINDING Ligands: PO4, GOL, K |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-05-25 Classification: HYDROLASE Ligands: ZN, GOL |

